
Methanolobus sp. SY-01
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanolobus; unclassified Methanolobus
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
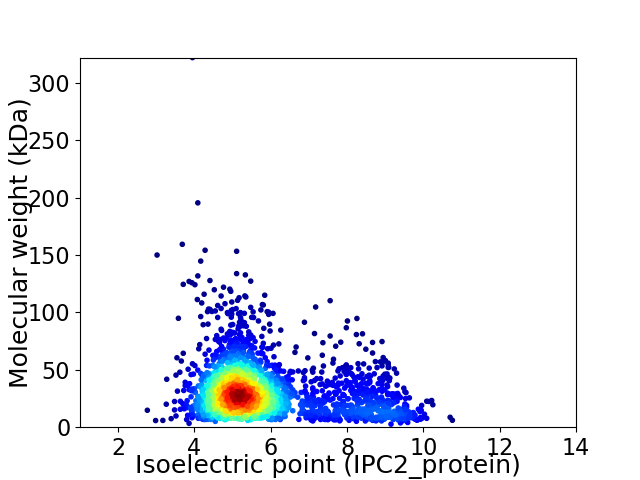
Virtual 2D-PAGE plot for 2452 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4E0PX16|A0A4E0PX16_9EURY Two-component system response regulator OS=Methanolobus sp. SY-01 OX=2052935 GN=CUN85_07250 PE=4 SV=1
MM1 pKa = 7.56ISKK4 pKa = 10.34KK5 pKa = 10.51SVILCFLSILLMASIDD21 pKa = 3.52ASIAQTIDD29 pKa = 2.86GTEE32 pKa = 3.66IQITSNRR39 pKa = 11.84SDD41 pKa = 3.1QYY43 pKa = 11.43FPAIYY48 pKa = 10.14DD49 pKa = 4.61DD50 pKa = 4.46IIVWTDD56 pKa = 2.84VRR58 pKa = 11.84YY59 pKa = 9.5EE60 pKa = 3.88SSDD63 pKa = 2.75IFLYY67 pKa = 11.16NMTSGEE73 pKa = 4.02EE74 pKa = 3.85LQITSNEE81 pKa = 4.07SEE83 pKa = 4.02EE84 pKa = 4.36RR85 pKa = 11.84VPDD88 pKa = 3.47IYY90 pKa = 11.38GDD92 pKa = 3.74IIVWEE97 pKa = 4.51DD98 pKa = 2.91WRR100 pKa = 11.84NGNWDD105 pKa = 3.25IYY107 pKa = 10.48IYY109 pKa = 11.0NLTSEE114 pKa = 4.7EE115 pKa = 3.9EE116 pKa = 4.16SALITNEE123 pKa = 3.2ADD125 pKa = 3.24QRR127 pKa = 11.84VPAIHH132 pKa = 7.37EE133 pKa = 4.07DD134 pKa = 3.77RR135 pKa = 11.84IVWEE139 pKa = 4.51DD140 pKa = 3.58LRR142 pKa = 11.84NGNWDD147 pKa = 2.71IYY149 pKa = 9.96MYY151 pKa = 10.26NISSGEE157 pKa = 3.87EE158 pKa = 3.65LQITDD163 pKa = 3.91NEE165 pKa = 4.36SDD167 pKa = 3.31QLYY170 pKa = 8.52PAVYY174 pKa = 9.92GDD176 pKa = 4.29LVTWMDD182 pKa = 4.1DD183 pKa = 3.8RR184 pKa = 11.84NGGWDD189 pKa = 3.07IFVYY193 pKa = 10.41DD194 pKa = 4.23LSSRR198 pKa = 11.84EE199 pKa = 3.99EE200 pKa = 3.99TQITANEE207 pKa = 4.36SIALYY212 pKa = 9.85PDD214 pKa = 3.48VYY216 pKa = 10.49EE217 pKa = 5.62DD218 pKa = 3.28RR219 pKa = 11.84VVWMDD224 pKa = 4.69DD225 pKa = 2.97RR226 pKa = 11.84NYY228 pKa = 10.59DD229 pKa = 3.86QNIFMYY235 pKa = 9.92NVSSEE240 pKa = 4.39EE241 pKa = 3.84EE242 pKa = 4.12LMVTTNEE249 pKa = 3.79SWQVVPKK256 pKa = 10.2IHH258 pKa = 7.42GDD260 pKa = 3.6RR261 pKa = 11.84IVWTDD266 pKa = 3.38EE267 pKa = 4.34RR268 pKa = 11.84NSNWDD273 pKa = 3.0ILGNSDD279 pKa = 3.17IYY281 pKa = 10.8MYY283 pKa = 10.07DD284 pKa = 3.0IPTGEE289 pKa = 4.29EE290 pKa = 3.8LQITTNEE297 pKa = 3.7EE298 pKa = 3.85RR299 pKa = 11.84QVYY302 pKa = 7.99PAIYY306 pKa = 9.31NDD308 pKa = 3.62TVVWEE313 pKa = 4.37DD314 pKa = 3.71RR315 pKa = 11.84RR316 pKa = 11.84NLNADD321 pKa = 3.46IYY323 pKa = 10.34IFTITGQNSTQQ334 pKa = 3.06
MM1 pKa = 7.56ISKK4 pKa = 10.34KK5 pKa = 10.51SVILCFLSILLMASIDD21 pKa = 3.52ASIAQTIDD29 pKa = 2.86GTEE32 pKa = 3.66IQITSNRR39 pKa = 11.84SDD41 pKa = 3.1QYY43 pKa = 11.43FPAIYY48 pKa = 10.14DD49 pKa = 4.61DD50 pKa = 4.46IIVWTDD56 pKa = 2.84VRR58 pKa = 11.84YY59 pKa = 9.5EE60 pKa = 3.88SSDD63 pKa = 2.75IFLYY67 pKa = 11.16NMTSGEE73 pKa = 4.02EE74 pKa = 3.85LQITSNEE81 pKa = 4.07SEE83 pKa = 4.02EE84 pKa = 4.36RR85 pKa = 11.84VPDD88 pKa = 3.47IYY90 pKa = 11.38GDD92 pKa = 3.74IIVWEE97 pKa = 4.51DD98 pKa = 2.91WRR100 pKa = 11.84NGNWDD105 pKa = 3.25IYY107 pKa = 10.48IYY109 pKa = 11.0NLTSEE114 pKa = 4.7EE115 pKa = 3.9EE116 pKa = 4.16SALITNEE123 pKa = 3.2ADD125 pKa = 3.24QRR127 pKa = 11.84VPAIHH132 pKa = 7.37EE133 pKa = 4.07DD134 pKa = 3.77RR135 pKa = 11.84IVWEE139 pKa = 4.51DD140 pKa = 3.58LRR142 pKa = 11.84NGNWDD147 pKa = 2.71IYY149 pKa = 9.96MYY151 pKa = 10.26NISSGEE157 pKa = 3.87EE158 pKa = 3.65LQITDD163 pKa = 3.91NEE165 pKa = 4.36SDD167 pKa = 3.31QLYY170 pKa = 8.52PAVYY174 pKa = 9.92GDD176 pKa = 4.29LVTWMDD182 pKa = 4.1DD183 pKa = 3.8RR184 pKa = 11.84NGGWDD189 pKa = 3.07IFVYY193 pKa = 10.41DD194 pKa = 4.23LSSRR198 pKa = 11.84EE199 pKa = 3.99EE200 pKa = 3.99TQITANEE207 pKa = 4.36SIALYY212 pKa = 9.85PDD214 pKa = 3.48VYY216 pKa = 10.49EE217 pKa = 5.62DD218 pKa = 3.28RR219 pKa = 11.84VVWMDD224 pKa = 4.69DD225 pKa = 2.97RR226 pKa = 11.84NYY228 pKa = 10.59DD229 pKa = 3.86QNIFMYY235 pKa = 9.92NVSSEE240 pKa = 4.39EE241 pKa = 3.84EE242 pKa = 4.12LMVTTNEE249 pKa = 3.79SWQVVPKK256 pKa = 10.2IHH258 pKa = 7.42GDD260 pKa = 3.6RR261 pKa = 11.84IVWTDD266 pKa = 3.38EE267 pKa = 4.34RR268 pKa = 11.84NSNWDD273 pKa = 3.0ILGNSDD279 pKa = 3.17IYY281 pKa = 10.8MYY283 pKa = 10.07DD284 pKa = 3.0IPTGEE289 pKa = 4.29EE290 pKa = 3.8LQITTNEE297 pKa = 3.7EE298 pKa = 3.85RR299 pKa = 11.84QVYY302 pKa = 7.99PAIYY306 pKa = 9.31NDD308 pKa = 3.62TVVWEE313 pKa = 4.37DD314 pKa = 3.71RR315 pKa = 11.84RR316 pKa = 11.84NLNADD321 pKa = 3.46IYY323 pKa = 10.34IFTITGQNSTQQ334 pKa = 3.06
Molecular weight: 39.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4E0QRM6|A0A4E0QRM6_9EURY DUF11 domain-containing protein OS=Methanolobus sp. SY-01 OX=2052935 GN=CUN85_07365 PE=4 SV=1
MM1 pKa = 7.14SHH3 pKa = 5.37NTKK6 pKa = 9.76GQKK9 pKa = 9.82ARR11 pKa = 11.84LAKK14 pKa = 10.24AHH16 pKa = 6.28RR17 pKa = 11.84QNQRR21 pKa = 11.84VPVWAIVKK29 pKa = 7.42TNRR32 pKa = 11.84KK33 pKa = 8.88VVGHH37 pKa = 6.0PKK39 pKa = 8.65RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.99WRR44 pKa = 11.84RR45 pKa = 11.84SSLDD49 pKa = 3.23VKK51 pKa = 11.09
MM1 pKa = 7.14SHH3 pKa = 5.37NTKK6 pKa = 9.76GQKK9 pKa = 9.82ARR11 pKa = 11.84LAKK14 pKa = 10.24AHH16 pKa = 6.28RR17 pKa = 11.84QNQRR21 pKa = 11.84VPVWAIVKK29 pKa = 7.42TNRR32 pKa = 11.84KK33 pKa = 8.88VVGHH37 pKa = 6.0PKK39 pKa = 8.65RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.99WRR44 pKa = 11.84RR45 pKa = 11.84SSLDD49 pKa = 3.23VKK51 pKa = 11.09
Molecular weight: 6.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
719907 |
25 |
2895 |
293.6 |
32.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.922 ± 0.053 | 1.222 ± 0.023 |
6.151 ± 0.05 | 7.487 ± 0.054 |
3.976 ± 0.036 | 7.216 ± 0.044 |
1.946 ± 0.023 | 8.127 ± 0.046 |
6.049 ± 0.046 | 8.965 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.996 ± 0.027 | 4.278 ± 0.045 |
3.787 ± 0.03 | 2.572 ± 0.022 |
4.729 ± 0.039 | 6.77 ± 0.049 |
5.346 ± 0.043 | 7.236 ± 0.036 |
0.843 ± 0.017 | 3.383 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
