
Desulfuribacillus alkaliarsenatis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillales incertae sedis; Desulfuribacillus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
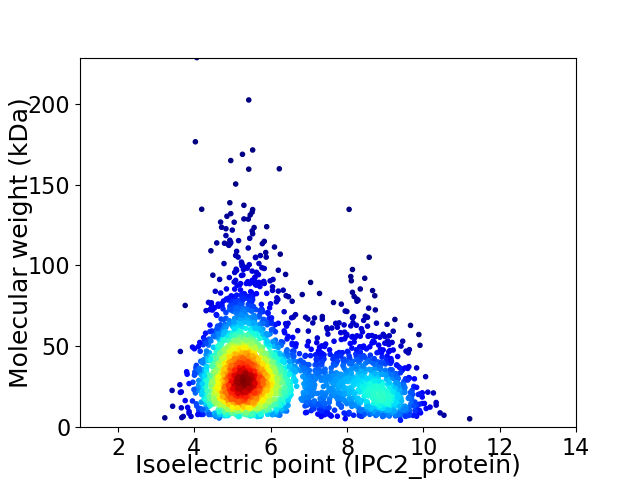
Virtual 2D-PAGE plot for 2879 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E5G3B0|A0A1E5G3B0_9BACL Uncharacterized protein OS=Desulfuribacillus alkaliarsenatis OX=766136 GN=BHF68_05585 PE=4 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84FTKK5 pKa = 10.18KK6 pKa = 10.13VVSICVIFMFILGTALPTGLAAEE29 pKa = 4.13QQIQINFNGQSYY41 pKa = 10.09NADD44 pKa = 3.92FYY46 pKa = 11.78LEE48 pKa = 5.16DD49 pKa = 6.1DD50 pKa = 3.14ITYY53 pKa = 10.56INVDD57 pKa = 3.85SLKK60 pKa = 10.36KK61 pKa = 10.39IPGLEE66 pKa = 4.03VNEE69 pKa = 4.1YY70 pKa = 10.79NYY72 pKa = 10.68VPLRR76 pKa = 11.84AFFEE80 pKa = 4.69GQGATVTWNAQSKK93 pKa = 10.61VIDD96 pKa = 3.71ISWRR100 pKa = 11.84EE101 pKa = 4.05TNDD104 pKa = 2.24GWTANDD110 pKa = 3.58LVVKK114 pKa = 10.69AEE116 pKa = 4.21EE117 pKa = 4.0VLKK120 pKa = 10.66EE121 pKa = 3.85YY122 pKa = 9.39NTYY125 pKa = 10.9RR126 pKa = 11.84MAGDD130 pKa = 3.49GTIDD134 pKa = 3.54ISISGNDD141 pKa = 3.49AADD144 pKa = 3.78AFPFDD149 pKa = 4.0GMTLAIEE156 pKa = 4.88GSFSQEE162 pKa = 3.42PLAMYY167 pKa = 7.52MKK169 pKa = 9.48QTMTMPINFEE179 pKa = 4.05EE180 pKa = 4.19MGLTEE185 pKa = 4.95EE186 pKa = 4.58EE187 pKa = 4.13LALMGMDD194 pKa = 3.1TSMVTEE200 pKa = 4.5MVWKK204 pKa = 10.65DD205 pKa = 3.1NAIYY209 pKa = 10.46QRR211 pKa = 11.84ILPLQEE217 pKa = 3.3QWIVNNIEE225 pKa = 4.22DD226 pKa = 4.69LGMSDD231 pKa = 3.85TFNDD235 pKa = 4.28LLQMTPQQNLEE246 pKa = 4.11LMTKK250 pKa = 10.16YY251 pKa = 10.74GVFYY255 pKa = 10.52TFGDD259 pKa = 3.85DD260 pKa = 3.91VEE262 pKa = 4.54VDD264 pKa = 3.82GIEE267 pKa = 4.33YY268 pKa = 10.62YY269 pKa = 10.7VVNSTLSADD278 pKa = 3.67TFRR281 pKa = 11.84NLMEE285 pKa = 5.99DD286 pKa = 3.25MFDD289 pKa = 4.51DD290 pKa = 4.96LMPPEE295 pKa = 5.0LLTEE299 pKa = 4.29EE300 pKa = 4.15SDD302 pKa = 3.8EE303 pKa = 4.31VSVMIDD309 pKa = 3.32EE310 pKa = 4.53IFANINMNFYY320 pKa = 10.54QQTLINKK327 pKa = 7.44EE328 pKa = 4.12TLTTDD333 pKa = 3.68FLIYY337 pKa = 10.94DD338 pKa = 3.91MVMSLALADD347 pKa = 3.98MFSQEE352 pKa = 5.89DD353 pKa = 4.33GLQEE357 pKa = 4.16DD358 pKa = 4.87APEE361 pKa = 4.24SMLMSMSGVYY371 pKa = 9.94HH372 pKa = 7.17LYY374 pKa = 10.89DD375 pKa = 3.7FGVEE379 pKa = 3.9IEE381 pKa = 4.82LPDD384 pKa = 3.48VSDD387 pKa = 5.62AITQEE392 pKa = 4.36EE393 pKa = 4.92YY394 pKa = 10.2MEE396 pKa = 3.92QQMQLFEE403 pKa = 4.34SLEE406 pKa = 4.21PEE408 pKa = 4.41VQQ410 pKa = 3.1
MM1 pKa = 7.98RR2 pKa = 11.84FTKK5 pKa = 10.18KK6 pKa = 10.13VVSICVIFMFILGTALPTGLAAEE29 pKa = 4.13QQIQINFNGQSYY41 pKa = 10.09NADD44 pKa = 3.92FYY46 pKa = 11.78LEE48 pKa = 5.16DD49 pKa = 6.1DD50 pKa = 3.14ITYY53 pKa = 10.56INVDD57 pKa = 3.85SLKK60 pKa = 10.36KK61 pKa = 10.39IPGLEE66 pKa = 4.03VNEE69 pKa = 4.1YY70 pKa = 10.79NYY72 pKa = 10.68VPLRR76 pKa = 11.84AFFEE80 pKa = 4.69GQGATVTWNAQSKK93 pKa = 10.61VIDD96 pKa = 3.71ISWRR100 pKa = 11.84EE101 pKa = 4.05TNDD104 pKa = 2.24GWTANDD110 pKa = 3.58LVVKK114 pKa = 10.69AEE116 pKa = 4.21EE117 pKa = 4.0VLKK120 pKa = 10.66EE121 pKa = 3.85YY122 pKa = 9.39NTYY125 pKa = 10.9RR126 pKa = 11.84MAGDD130 pKa = 3.49GTIDD134 pKa = 3.54ISISGNDD141 pKa = 3.49AADD144 pKa = 3.78AFPFDD149 pKa = 4.0GMTLAIEE156 pKa = 4.88GSFSQEE162 pKa = 3.42PLAMYY167 pKa = 7.52MKK169 pKa = 9.48QTMTMPINFEE179 pKa = 4.05EE180 pKa = 4.19MGLTEE185 pKa = 4.95EE186 pKa = 4.58EE187 pKa = 4.13LALMGMDD194 pKa = 3.1TSMVTEE200 pKa = 4.5MVWKK204 pKa = 10.65DD205 pKa = 3.1NAIYY209 pKa = 10.46QRR211 pKa = 11.84ILPLQEE217 pKa = 3.3QWIVNNIEE225 pKa = 4.22DD226 pKa = 4.69LGMSDD231 pKa = 3.85TFNDD235 pKa = 4.28LLQMTPQQNLEE246 pKa = 4.11LMTKK250 pKa = 10.16YY251 pKa = 10.74GVFYY255 pKa = 10.52TFGDD259 pKa = 3.85DD260 pKa = 3.91VEE262 pKa = 4.54VDD264 pKa = 3.82GIEE267 pKa = 4.33YY268 pKa = 10.62YY269 pKa = 10.7VVNSTLSADD278 pKa = 3.67TFRR281 pKa = 11.84NLMEE285 pKa = 5.99DD286 pKa = 3.25MFDD289 pKa = 4.51DD290 pKa = 4.96LMPPEE295 pKa = 5.0LLTEE299 pKa = 4.29EE300 pKa = 4.15SDD302 pKa = 3.8EE303 pKa = 4.31VSVMIDD309 pKa = 3.32EE310 pKa = 4.53IFANINMNFYY320 pKa = 10.54QQTLINKK327 pKa = 7.44EE328 pKa = 4.12TLTTDD333 pKa = 3.68FLIYY337 pKa = 10.94DD338 pKa = 3.91MVMSLALADD347 pKa = 3.98MFSQEE352 pKa = 5.89DD353 pKa = 4.33GLQEE357 pKa = 4.16DD358 pKa = 4.87APEE361 pKa = 4.24SMLMSMSGVYY371 pKa = 9.94HH372 pKa = 7.17LYY374 pKa = 10.89DD375 pKa = 3.7FGVEE379 pKa = 3.9IEE381 pKa = 4.82LPDD384 pKa = 3.48VSDD387 pKa = 5.62AITQEE392 pKa = 4.36EE393 pKa = 4.92YY394 pKa = 10.2MEE396 pKa = 3.92QQMQLFEE403 pKa = 4.34SLEE406 pKa = 4.21PEE408 pKa = 4.41VQQ410 pKa = 3.1
Molecular weight: 46.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E5G318|A0A1E5G318_9BACL Arsenic-transporting ATPase OS=Desulfuribacillus alkaliarsenatis OX=766136 GN=BHF68_04430 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTRR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84KK13 pKa = 8.71KK14 pKa = 8.74VHH16 pKa = 5.61GFRR19 pKa = 11.84KK20 pKa = 9.89RR21 pKa = 11.84MSSSSGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.38KK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.71VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTRR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84KK13 pKa = 8.71KK14 pKa = 8.74VHH16 pKa = 5.61GFRR19 pKa = 11.84KK20 pKa = 9.89RR21 pKa = 11.84MSSSSGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.38KK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.71VLSAA44 pKa = 4.05
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
915468 |
37 |
2151 |
318.0 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.119 ± 0.047 | 0.867 ± 0.019 |
5.354 ± 0.036 | 7.215 ± 0.049 |
4.078 ± 0.033 | 6.485 ± 0.042 |
1.94 ± 0.021 | 8.811 ± 0.047 |
6.574 ± 0.052 | 9.604 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.636 ± 0.022 | 4.895 ± 0.038 |
3.373 ± 0.022 | 3.985 ± 0.035 |
4.213 ± 0.032 | 5.807 ± 0.029 |
5.366 ± 0.028 | 7.074 ± 0.036 |
0.886 ± 0.015 | 3.719 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
