
Avon-Heathcote Estuary associated circular virus 24
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.75
Get precalculated fractions of proteins
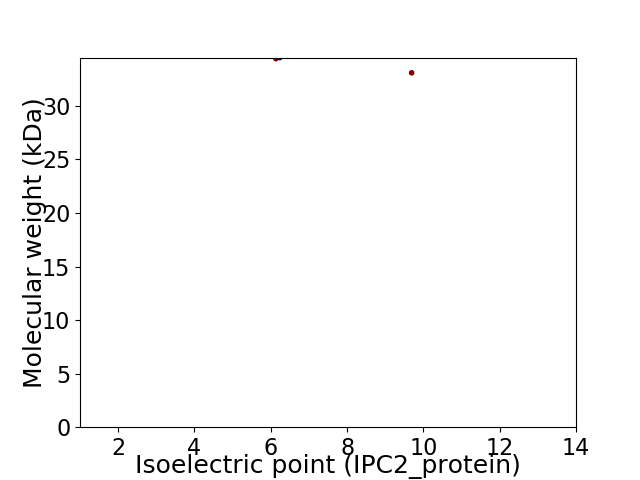
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IB82|A0A0C5IB82_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 24 OX=1618248 PE=4 SV=1
MM1 pKa = 7.04AQSRR5 pKa = 11.84SWCFTLNNYY14 pKa = 7.96VQADD18 pKa = 3.79IDD20 pKa = 3.6RR21 pKa = 11.84LAAFGEE27 pKa = 4.5TDD29 pKa = 3.28DD30 pKa = 3.9CTYY33 pKa = 11.26LVFGKK38 pKa = 10.34EE39 pKa = 4.14VGEE42 pKa = 4.34SGTPHH47 pKa = 6.17LQGFAIFPRR56 pKa = 11.84KK57 pKa = 9.44LRR59 pKa = 11.84LRR61 pKa = 11.84AVKK64 pKa = 10.13SHH66 pKa = 6.75IGNGAHH72 pKa = 6.93LEE74 pKa = 4.19CARR77 pKa = 11.84GSPLQASDD85 pKa = 3.65YY86 pKa = 10.15CKK88 pKa = 10.49KK89 pKa = 10.86DD90 pKa = 2.89GDD92 pKa = 3.88FTEE95 pKa = 5.15FGSFGGVRR103 pKa = 11.84PGLSGRR109 pKa = 11.84FGQFVEE115 pKa = 4.62WLDD118 pKa = 3.45EE119 pKa = 4.32HH120 pKa = 7.04YY121 pKa = 11.0KK122 pKa = 10.29EE123 pKa = 5.27GNVHH127 pKa = 5.33QPSRR131 pKa = 11.84ALIAATWPDD140 pKa = 3.61LYY142 pKa = 11.33VRR144 pKa = 11.84YY145 pKa = 8.32HH146 pKa = 5.87TKK148 pKa = 10.45LFEE151 pKa = 4.34LVGVLAPKK159 pKa = 10.27PILQEE164 pKa = 3.89GDD166 pKa = 3.16PNAWQTTLIEE176 pKa = 4.5ALGLDD181 pKa = 3.56PDD183 pKa = 3.68DD184 pKa = 4.49RR185 pKa = 11.84KK186 pKa = 10.64IYY188 pKa = 10.2FYY190 pKa = 11.39VDD192 pKa = 3.07EE193 pKa = 4.94TGGSGKK199 pKa = 10.07SWLTRR204 pKa = 11.84YY205 pKa = 10.52LMTSRR210 pKa = 11.84DD211 pKa = 3.77DD212 pKa = 3.69VQALSIGKK220 pKa = 9.51RR221 pKa = 11.84DD222 pKa = 4.82DD223 pKa = 3.07IAHH226 pKa = 7.1AIDD229 pKa = 3.48VTKK232 pKa = 10.62KK233 pKa = 9.76VFLFNVPRR241 pKa = 11.84TQMEE245 pKa = 4.25FLQYY249 pKa = 10.86SILEE253 pKa = 4.2SLKK256 pKa = 10.54DD257 pKa = 3.68RR258 pKa = 11.84MVMSPKK264 pKa = 9.37YY265 pKa = 9.59NSMMKK270 pKa = 9.94VLHH273 pKa = 6.07SVPHH277 pKa = 5.07VVVFSNEE284 pKa = 3.75EE285 pKa = 3.67PDD287 pKa = 3.73RR288 pKa = 11.84TKK290 pKa = 10.55LTADD294 pKa = 3.88RR295 pKa = 11.84FAVTHH300 pKa = 5.9IRR302 pKa = 11.84NII304 pKa = 3.59
MM1 pKa = 7.04AQSRR5 pKa = 11.84SWCFTLNNYY14 pKa = 7.96VQADD18 pKa = 3.79IDD20 pKa = 3.6RR21 pKa = 11.84LAAFGEE27 pKa = 4.5TDD29 pKa = 3.28DD30 pKa = 3.9CTYY33 pKa = 11.26LVFGKK38 pKa = 10.34EE39 pKa = 4.14VGEE42 pKa = 4.34SGTPHH47 pKa = 6.17LQGFAIFPRR56 pKa = 11.84KK57 pKa = 9.44LRR59 pKa = 11.84LRR61 pKa = 11.84AVKK64 pKa = 10.13SHH66 pKa = 6.75IGNGAHH72 pKa = 6.93LEE74 pKa = 4.19CARR77 pKa = 11.84GSPLQASDD85 pKa = 3.65YY86 pKa = 10.15CKK88 pKa = 10.49KK89 pKa = 10.86DD90 pKa = 2.89GDD92 pKa = 3.88FTEE95 pKa = 5.15FGSFGGVRR103 pKa = 11.84PGLSGRR109 pKa = 11.84FGQFVEE115 pKa = 4.62WLDD118 pKa = 3.45EE119 pKa = 4.32HH120 pKa = 7.04YY121 pKa = 11.0KK122 pKa = 10.29EE123 pKa = 5.27GNVHH127 pKa = 5.33QPSRR131 pKa = 11.84ALIAATWPDD140 pKa = 3.61LYY142 pKa = 11.33VRR144 pKa = 11.84YY145 pKa = 8.32HH146 pKa = 5.87TKK148 pKa = 10.45LFEE151 pKa = 4.34LVGVLAPKK159 pKa = 10.27PILQEE164 pKa = 3.89GDD166 pKa = 3.16PNAWQTTLIEE176 pKa = 4.5ALGLDD181 pKa = 3.56PDD183 pKa = 3.68DD184 pKa = 4.49RR185 pKa = 11.84KK186 pKa = 10.64IYY188 pKa = 10.2FYY190 pKa = 11.39VDD192 pKa = 3.07EE193 pKa = 4.94TGGSGKK199 pKa = 10.07SWLTRR204 pKa = 11.84YY205 pKa = 10.52LMTSRR210 pKa = 11.84DD211 pKa = 3.77DD212 pKa = 3.69VQALSIGKK220 pKa = 9.51RR221 pKa = 11.84DD222 pKa = 4.82DD223 pKa = 3.07IAHH226 pKa = 7.1AIDD229 pKa = 3.48VTKK232 pKa = 10.62KK233 pKa = 9.76VFLFNVPRR241 pKa = 11.84TQMEE245 pKa = 4.25FLQYY249 pKa = 10.86SILEE253 pKa = 4.2SLKK256 pKa = 10.54DD257 pKa = 3.68RR258 pKa = 11.84MVMSPKK264 pKa = 9.37YY265 pKa = 9.59NSMMKK270 pKa = 9.94VLHH273 pKa = 6.07SVPHH277 pKa = 5.07VVVFSNEE284 pKa = 3.75EE285 pKa = 3.67PDD287 pKa = 3.73RR288 pKa = 11.84TKK290 pKa = 10.55LTADD294 pKa = 3.88RR295 pKa = 11.84FAVTHH300 pKa = 5.9IRR302 pKa = 11.84NII304 pKa = 3.59
Molecular weight: 34.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IB82|A0A0C5IB82_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 24 OX=1618248 PE=4 SV=1
MM1 pKa = 7.19SAVVTYY7 pKa = 9.64GPRR10 pKa = 11.84AARR13 pKa = 11.84FAMRR17 pKa = 11.84TYY19 pKa = 10.16RR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84VATYY28 pKa = 10.84ARR30 pKa = 11.84GARR33 pKa = 11.84MALNLARR40 pKa = 11.84NKK42 pKa = 10.03RR43 pKa = 11.84VRR45 pKa = 11.84QAVGQAYY52 pKa = 9.87RR53 pKa = 11.84RR54 pKa = 11.84VKK56 pKa = 10.12RR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 7.27TNSFKK64 pKa = 10.91GRR66 pKa = 11.84APRR69 pKa = 11.84SNKK72 pKa = 8.86QRR74 pKa = 11.84SNTQNNQQLVVNHH87 pKa = 6.86LEE89 pKa = 4.06STIPSWPPAASDD101 pKa = 3.75SDD103 pKa = 3.77PKK105 pKa = 10.91QGFRR109 pKa = 11.84RR110 pKa = 11.84GNFIMLDD117 pKa = 3.45ALKK120 pKa = 10.94GCWTFKK126 pKa = 10.69NSGRR130 pKa = 11.84YY131 pKa = 8.12PVTVHH136 pKa = 6.25FACVQSVADD145 pKa = 4.1ANANAKK151 pKa = 9.57IDD153 pKa = 3.88FFSNPGGTDD162 pKa = 2.82KK163 pKa = 11.63YY164 pKa = 11.28KK165 pKa = 11.25DD166 pKa = 3.66FVDD169 pKa = 3.77SKK171 pKa = 11.58NVFDD175 pKa = 5.45IDD177 pKa = 4.51KK178 pKa = 10.97DD179 pKa = 3.99CLGLNKK185 pKa = 10.43NKK187 pKa = 10.17FNILFHH193 pKa = 6.32KK194 pKa = 10.21KK195 pKa = 9.39IYY197 pKa = 10.24LEE199 pKa = 4.29EE200 pKa = 5.42GEE202 pKa = 4.98HH203 pKa = 6.69VDD205 pKa = 3.38GANIRR210 pKa = 11.84TYY212 pKa = 11.01SLSEE216 pKa = 3.94SKK218 pKa = 10.93YY219 pKa = 7.82MKK221 pKa = 9.55TIRR224 pKa = 11.84VYY226 pKa = 10.8RR227 pKa = 11.84PIKK230 pKa = 10.43KK231 pKa = 10.33NIEE234 pKa = 4.13FEE236 pKa = 4.33NKK238 pKa = 8.5STLEE242 pKa = 3.61PLKK245 pKa = 10.6NIKK248 pKa = 9.41YY249 pKa = 9.77LYY251 pKa = 9.21WFVPRR256 pKa = 11.84YY257 pKa = 8.98STDD260 pKa = 3.1QPLAGQTKK268 pKa = 8.74GEE270 pKa = 4.01RR271 pKa = 11.84AVTVDD276 pKa = 3.05HH277 pKa = 6.97TDD279 pKa = 2.92SVLYY283 pKa = 10.06RR284 pKa = 11.84DD285 pKa = 3.98RR286 pKa = 11.84LL287 pKa = 3.62
MM1 pKa = 7.19SAVVTYY7 pKa = 9.64GPRR10 pKa = 11.84AARR13 pKa = 11.84FAMRR17 pKa = 11.84TYY19 pKa = 10.16RR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84VATYY28 pKa = 10.84ARR30 pKa = 11.84GARR33 pKa = 11.84MALNLARR40 pKa = 11.84NKK42 pKa = 10.03RR43 pKa = 11.84VRR45 pKa = 11.84QAVGQAYY52 pKa = 9.87RR53 pKa = 11.84RR54 pKa = 11.84VKK56 pKa = 10.12RR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 7.27TNSFKK64 pKa = 10.91GRR66 pKa = 11.84APRR69 pKa = 11.84SNKK72 pKa = 8.86QRR74 pKa = 11.84SNTQNNQQLVVNHH87 pKa = 6.86LEE89 pKa = 4.06STIPSWPPAASDD101 pKa = 3.75SDD103 pKa = 3.77PKK105 pKa = 10.91QGFRR109 pKa = 11.84RR110 pKa = 11.84GNFIMLDD117 pKa = 3.45ALKK120 pKa = 10.94GCWTFKK126 pKa = 10.69NSGRR130 pKa = 11.84YY131 pKa = 8.12PVTVHH136 pKa = 6.25FACVQSVADD145 pKa = 4.1ANANAKK151 pKa = 9.57IDD153 pKa = 3.88FFSNPGGTDD162 pKa = 2.82KK163 pKa = 11.63YY164 pKa = 11.28KK165 pKa = 11.25DD166 pKa = 3.66FVDD169 pKa = 3.77SKK171 pKa = 11.58NVFDD175 pKa = 5.45IDD177 pKa = 4.51KK178 pKa = 10.97DD179 pKa = 3.99CLGLNKK185 pKa = 10.43NKK187 pKa = 10.17FNILFHH193 pKa = 6.32KK194 pKa = 10.21KK195 pKa = 9.39IYY197 pKa = 10.24LEE199 pKa = 4.29EE200 pKa = 5.42GEE202 pKa = 4.98HH203 pKa = 6.69VDD205 pKa = 3.38GANIRR210 pKa = 11.84TYY212 pKa = 11.01SLSEE216 pKa = 3.94SKK218 pKa = 10.93YY219 pKa = 7.82MKK221 pKa = 9.55TIRR224 pKa = 11.84VYY226 pKa = 10.8RR227 pKa = 11.84PIKK230 pKa = 10.43KK231 pKa = 10.33NIEE234 pKa = 4.13FEE236 pKa = 4.33NKK238 pKa = 8.5STLEE242 pKa = 3.61PLKK245 pKa = 10.6NIKK248 pKa = 9.41YY249 pKa = 9.77LYY251 pKa = 9.21WFVPRR256 pKa = 11.84YY257 pKa = 8.98STDD260 pKa = 3.1QPLAGQTKK268 pKa = 8.74GEE270 pKa = 4.01RR271 pKa = 11.84AVTVDD276 pKa = 3.05HH277 pKa = 6.97TDD279 pKa = 2.92SVLYY283 pKa = 10.06RR284 pKa = 11.84DD285 pKa = 3.98RR286 pKa = 11.84LL287 pKa = 3.62
Molecular weight: 33.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
591 |
287 |
304 |
295.5 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.276 ± 0.468 | 1.184 ± 0.088 |
6.43 ± 0.542 | 4.23 ± 0.694 |
5.245 ± 0.233 | 6.43 ± 0.763 |
2.538 ± 0.505 | 4.061 ± 0.145 |
7.107 ± 1.018 | 7.614 ± 1.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.183 | 5.245 ± 1.536 |
4.399 ± 0.138 | 3.553 ± 0.044 |
8.122 ± 1.258 | 6.261 ± 0.007 |
5.584 ± 0.006 | 7.107 ± 0.088 |
1.354 ± 0.196 | 4.23 ± 0.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
