
Psychroflexus sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Psychroflexus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
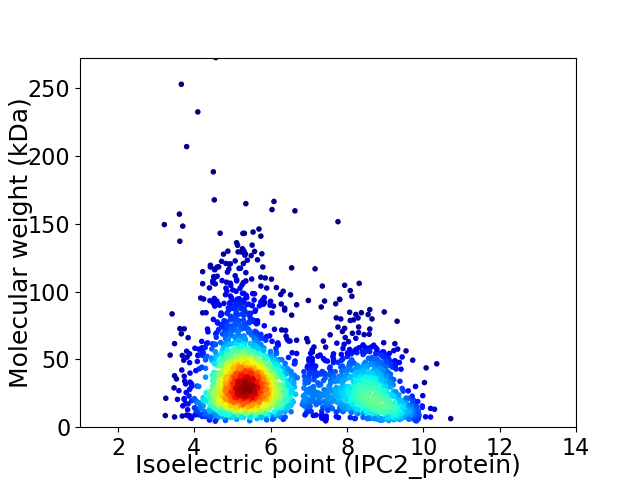
Virtual 2D-PAGE plot for 2655 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7XD03|A0A1G7XD03_9FLAO mRNA interferase HigB OS=Psychroflexus sediminis OX=470826 GN=SAMN04488027_10833 PE=4 SV=1
MM1 pKa = 6.91KK2 pKa = 10.22QIYY5 pKa = 9.91SLLILFIVSLSYY17 pKa = 11.04GQVINPGTTSANIVSNSAPNITCPEE42 pKa = 4.58EE43 pKa = 3.83ITTTATSPIVNFNNPTLTDD62 pKa = 3.5NEE64 pKa = 4.27TTAFPFVSSDD74 pKa = 3.05MTYY77 pKa = 10.15IGSNGGKK84 pKa = 9.57NFFISEE90 pKa = 4.05QSFSGANAFADD101 pKa = 5.0AIARR105 pKa = 11.84GGLVATISDD114 pKa = 3.54ATQNTFIANALQANNITEE132 pKa = 3.98AHH134 pKa = 6.32IGYY137 pKa = 9.75SDD139 pKa = 3.74AASEE143 pKa = 5.24GNFVWQDD150 pKa = 3.09GSTSGYY156 pKa = 10.16EE157 pKa = 3.48NWQPNEE163 pKa = 4.09PNDD166 pKa = 3.69TDD168 pKa = 3.59GGEE171 pKa = 4.35DD172 pKa = 4.04YY173 pKa = 10.67VTISAGGGWNDD184 pKa = 2.87VGARR188 pKa = 11.84GTGNARR194 pKa = 11.84YY195 pKa = 9.32IFQLDD200 pKa = 3.39QTDD203 pKa = 3.43GFLIQTSGLASGSSFPIGTTTNTFEE228 pKa = 5.75AYY230 pKa = 9.23DD231 pKa = 3.5ASGNSSICSFNVTVTDD247 pKa = 3.24IPEE250 pKa = 4.13VVTQNIDD257 pKa = 3.32VVLDD261 pKa = 3.41ATGNATISPEE271 pKa = 4.14EE272 pKa = 4.03IDD274 pKa = 3.64NGSTAVSGILGLSLDD289 pKa = 4.48ILNFDD294 pKa = 4.63CSNLGEE300 pKa = 4.26NTVTLTVISNTVEE313 pKa = 4.2TATGTAIVTVRR324 pKa = 11.84DD325 pKa = 3.76TTAPTVVTQDD335 pKa = 2.82ITVEE339 pKa = 3.96LDD341 pKa = 3.05ASGEE345 pKa = 4.17AIITADD351 pKa = 4.04QIDD354 pKa = 4.31NGSADD359 pKa = 3.42NCGIASLDD367 pKa = 3.58LSRR370 pKa = 11.84TNFNGFDD377 pKa = 3.01IGQNTVEE384 pKa = 4.59LKK386 pKa = 9.63VTDD389 pKa = 3.49SSGNQSAGTATVTIVDD405 pKa = 4.1TTPPNAVCSNQTIFLDD421 pKa = 3.75EE422 pKa = 4.75SGNASISASDD432 pKa = 3.59VDD434 pKa = 5.41GGSSDD439 pKa = 3.71NASAAYY445 pKa = 9.39IPLLNYY451 pKa = 10.11EE452 pKa = 4.61PIEE455 pKa = 4.36TGSGINASEE464 pKa = 4.42FEE466 pKa = 4.26FVAQVVTIPQPISDD480 pKa = 3.88FRR482 pKa = 11.84IKK484 pKa = 10.96LEE486 pKa = 4.29LATCKK491 pKa = 10.91SNGLTGATLDD501 pKa = 3.85IVEE504 pKa = 5.53LDD506 pKa = 3.89PSGHH510 pKa = 7.1PDD512 pKa = 3.13LSLVLGSSSVPEE524 pKa = 3.7SDD526 pKa = 2.84IYY528 pKa = 11.15GWPGCVSGSFGLTTFDD544 pKa = 4.1FSGVDD549 pKa = 3.6LQQGNQYY556 pKa = 11.66ALVLKK561 pKa = 10.32NSEE564 pKa = 4.11GARR567 pKa = 11.84GSVSWNRR574 pKa = 11.84SAIPTDD580 pKa = 3.48PSNPYY585 pKa = 9.82QGGNIWTYY593 pKa = 8.8EE594 pKa = 4.02NYY596 pKa = 10.11RR597 pKa = 11.84SRR599 pKa = 11.84LLSDD603 pKa = 3.44EE604 pKa = 4.12VWRR607 pKa = 11.84EE608 pKa = 4.0DD609 pKa = 3.59EE610 pKa = 4.11THH612 pKa = 7.14DD613 pKa = 3.52LHH615 pKa = 8.5FSIEE619 pKa = 3.99ALQEE623 pKa = 4.16DD624 pKa = 4.74LTYY627 pKa = 10.92SIDD630 pKa = 3.74QDD632 pKa = 4.74SFDD635 pKa = 4.28CSNLGVNTVSLTVTDD650 pKa = 4.11ASGNTSSCISEE661 pKa = 3.97VTVIDD666 pKa = 3.64NLAPTVVTQDD676 pKa = 2.82ITVEE680 pKa = 3.79LDD682 pKa = 3.21ANGEE686 pKa = 4.07AGITADD692 pKa = 4.51QIDD695 pKa = 4.45NGTSDD700 pKa = 3.67NCSSISGYY708 pKa = 10.31CIEE711 pKa = 4.6GMTYY715 pKa = 10.71GNAEE719 pKa = 3.93IFIEE723 pKa = 4.56GAGMGRR729 pKa = 11.84VDD731 pKa = 4.84FSAGLVSASSGGVPMEE747 pKa = 4.5LVPITTIDD755 pKa = 3.46SKK757 pKa = 11.41VRR759 pKa = 11.84SGDD762 pKa = 3.46LVHH765 pKa = 7.17ARR767 pKa = 11.84STLSTGSRR775 pKa = 11.84PSGEE779 pKa = 4.24SIFYY783 pKa = 10.0RR784 pKa = 11.84VSDD787 pKa = 3.99GRR789 pKa = 11.84IIWGGHH795 pKa = 4.58SPSGSFANRR804 pKa = 11.84TQLVIEE810 pKa = 4.3QPNVATGEE818 pKa = 4.46LIDD821 pKa = 4.55FSKK824 pKa = 10.68PYY826 pKa = 10.3NLKK829 pKa = 9.25FQLNGRR835 pKa = 11.84YY836 pKa = 9.12LSQSGGSFTSVSSPADD852 pKa = 3.06ISAVLISGDD861 pKa = 3.7LGATNNCLLTTSLDD875 pKa = 3.34ATTFDD880 pKa = 5.04CSNLGDD886 pKa = 3.91NEE888 pKa = 4.18VTLTVTDD895 pKa = 4.41GSGNSSSATAIVTVIDD911 pKa = 4.51NIAPTIVLKK920 pKa = 10.96DD921 pKa = 3.33VTVEE925 pKa = 3.89LNAEE929 pKa = 4.14GTVSVEE935 pKa = 4.59ASAFDD940 pKa = 3.53NGSFDD945 pKa = 3.41NCGEE949 pKa = 4.31VTFSTNLVEE958 pKa = 5.04ILSCGALGLNEE969 pKa = 4.07VTVTVTDD976 pKa = 3.69ASGNQSQNTAILTVLDD992 pKa = 4.08NLSPAVVTQDD1002 pKa = 3.11FTVVLNANGEE1012 pKa = 4.21ATITAAQVDD1021 pKa = 4.2NGSADD1026 pKa = 3.33NCGVDD1031 pKa = 4.04TLALDD1036 pKa = 3.5ITTFDD1041 pKa = 4.27CSNLGEE1047 pKa = 4.23NTVTLTATDD1056 pKa = 4.1GSGKK1060 pKa = 9.72QATATAIVTVVDD1072 pKa = 3.65NSLPTAVAQNLTVEE1086 pKa = 4.18LDD1088 pKa = 3.53EE1089 pKa = 5.56FGAASITAAQVDD1101 pKa = 4.03NGSSDD1106 pKa = 3.33NCGVEE1111 pKa = 4.31TLALDD1116 pKa = 3.77VTSFDD1121 pKa = 4.49CSNLGEE1127 pKa = 4.3NTVTLRR1133 pKa = 11.84VTDD1136 pKa = 4.12GSGNQATSTAVITVIDD1152 pKa = 4.21ALAPALVTADD1162 pKa = 3.13ITVSLDD1168 pKa = 3.21EE1169 pKa = 4.52NATVSIEE1176 pKa = 3.91PGNLVDD1182 pKa = 4.64TLEE1185 pKa = 5.03DD1186 pKa = 3.52NCSLAGEE1193 pKa = 4.25ITLALDD1199 pKa = 3.17QDD1201 pKa = 4.2TFTATGIYY1209 pKa = 9.25VVNITATDD1217 pKa = 3.41ASGNTTTEE1225 pKa = 4.17SAEE1228 pKa = 4.08VTVDD1232 pKa = 3.2STLSIDD1238 pKa = 4.4QVDD1241 pKa = 3.46SDD1243 pKa = 4.5IQVKK1247 pKa = 10.01LYY1249 pKa = 9.6PNPTTDD1255 pKa = 3.02HH1256 pKa = 6.76LFFEE1260 pKa = 4.9VSNTEE1265 pKa = 4.45LIHH1268 pKa = 5.15ITIYY1272 pKa = 11.35DD1273 pKa = 3.72MNGKK1277 pKa = 8.77LVKK1280 pKa = 9.85TSQEE1284 pKa = 3.94SHH1286 pKa = 6.22SVDD1289 pKa = 3.36VTNLSSGVYY1298 pKa = 8.62FAKK1301 pKa = 10.51VDD1303 pKa = 3.66AKK1305 pKa = 11.0GGNTLKK1311 pKa = 10.57ILRR1314 pKa = 11.84FIKK1317 pKa = 10.44KK1318 pKa = 9.73
MM1 pKa = 6.91KK2 pKa = 10.22QIYY5 pKa = 9.91SLLILFIVSLSYY17 pKa = 11.04GQVINPGTTSANIVSNSAPNITCPEE42 pKa = 4.58EE43 pKa = 3.83ITTTATSPIVNFNNPTLTDD62 pKa = 3.5NEE64 pKa = 4.27TTAFPFVSSDD74 pKa = 3.05MTYY77 pKa = 10.15IGSNGGKK84 pKa = 9.57NFFISEE90 pKa = 4.05QSFSGANAFADD101 pKa = 5.0AIARR105 pKa = 11.84GGLVATISDD114 pKa = 3.54ATQNTFIANALQANNITEE132 pKa = 3.98AHH134 pKa = 6.32IGYY137 pKa = 9.75SDD139 pKa = 3.74AASEE143 pKa = 5.24GNFVWQDD150 pKa = 3.09GSTSGYY156 pKa = 10.16EE157 pKa = 3.48NWQPNEE163 pKa = 4.09PNDD166 pKa = 3.69TDD168 pKa = 3.59GGEE171 pKa = 4.35DD172 pKa = 4.04YY173 pKa = 10.67VTISAGGGWNDD184 pKa = 2.87VGARR188 pKa = 11.84GTGNARR194 pKa = 11.84YY195 pKa = 9.32IFQLDD200 pKa = 3.39QTDD203 pKa = 3.43GFLIQTSGLASGSSFPIGTTTNTFEE228 pKa = 5.75AYY230 pKa = 9.23DD231 pKa = 3.5ASGNSSICSFNVTVTDD247 pKa = 3.24IPEE250 pKa = 4.13VVTQNIDD257 pKa = 3.32VVLDD261 pKa = 3.41ATGNATISPEE271 pKa = 4.14EE272 pKa = 4.03IDD274 pKa = 3.64NGSTAVSGILGLSLDD289 pKa = 4.48ILNFDD294 pKa = 4.63CSNLGEE300 pKa = 4.26NTVTLTVISNTVEE313 pKa = 4.2TATGTAIVTVRR324 pKa = 11.84DD325 pKa = 3.76TTAPTVVTQDD335 pKa = 2.82ITVEE339 pKa = 3.96LDD341 pKa = 3.05ASGEE345 pKa = 4.17AIITADD351 pKa = 4.04QIDD354 pKa = 4.31NGSADD359 pKa = 3.42NCGIASLDD367 pKa = 3.58LSRR370 pKa = 11.84TNFNGFDD377 pKa = 3.01IGQNTVEE384 pKa = 4.59LKK386 pKa = 9.63VTDD389 pKa = 3.49SSGNQSAGTATVTIVDD405 pKa = 4.1TTPPNAVCSNQTIFLDD421 pKa = 3.75EE422 pKa = 4.75SGNASISASDD432 pKa = 3.59VDD434 pKa = 5.41GGSSDD439 pKa = 3.71NASAAYY445 pKa = 9.39IPLLNYY451 pKa = 10.11EE452 pKa = 4.61PIEE455 pKa = 4.36TGSGINASEE464 pKa = 4.42FEE466 pKa = 4.26FVAQVVTIPQPISDD480 pKa = 3.88FRR482 pKa = 11.84IKK484 pKa = 10.96LEE486 pKa = 4.29LATCKK491 pKa = 10.91SNGLTGATLDD501 pKa = 3.85IVEE504 pKa = 5.53LDD506 pKa = 3.89PSGHH510 pKa = 7.1PDD512 pKa = 3.13LSLVLGSSSVPEE524 pKa = 3.7SDD526 pKa = 2.84IYY528 pKa = 11.15GWPGCVSGSFGLTTFDD544 pKa = 4.1FSGVDD549 pKa = 3.6LQQGNQYY556 pKa = 11.66ALVLKK561 pKa = 10.32NSEE564 pKa = 4.11GARR567 pKa = 11.84GSVSWNRR574 pKa = 11.84SAIPTDD580 pKa = 3.48PSNPYY585 pKa = 9.82QGGNIWTYY593 pKa = 8.8EE594 pKa = 4.02NYY596 pKa = 10.11RR597 pKa = 11.84SRR599 pKa = 11.84LLSDD603 pKa = 3.44EE604 pKa = 4.12VWRR607 pKa = 11.84EE608 pKa = 4.0DD609 pKa = 3.59EE610 pKa = 4.11THH612 pKa = 7.14DD613 pKa = 3.52LHH615 pKa = 8.5FSIEE619 pKa = 3.99ALQEE623 pKa = 4.16DD624 pKa = 4.74LTYY627 pKa = 10.92SIDD630 pKa = 3.74QDD632 pKa = 4.74SFDD635 pKa = 4.28CSNLGVNTVSLTVTDD650 pKa = 4.11ASGNTSSCISEE661 pKa = 3.97VTVIDD666 pKa = 3.64NLAPTVVTQDD676 pKa = 2.82ITVEE680 pKa = 3.79LDD682 pKa = 3.21ANGEE686 pKa = 4.07AGITADD692 pKa = 4.51QIDD695 pKa = 4.45NGTSDD700 pKa = 3.67NCSSISGYY708 pKa = 10.31CIEE711 pKa = 4.6GMTYY715 pKa = 10.71GNAEE719 pKa = 3.93IFIEE723 pKa = 4.56GAGMGRR729 pKa = 11.84VDD731 pKa = 4.84FSAGLVSASSGGVPMEE747 pKa = 4.5LVPITTIDD755 pKa = 3.46SKK757 pKa = 11.41VRR759 pKa = 11.84SGDD762 pKa = 3.46LVHH765 pKa = 7.17ARR767 pKa = 11.84STLSTGSRR775 pKa = 11.84PSGEE779 pKa = 4.24SIFYY783 pKa = 10.0RR784 pKa = 11.84VSDD787 pKa = 3.99GRR789 pKa = 11.84IIWGGHH795 pKa = 4.58SPSGSFANRR804 pKa = 11.84TQLVIEE810 pKa = 4.3QPNVATGEE818 pKa = 4.46LIDD821 pKa = 4.55FSKK824 pKa = 10.68PYY826 pKa = 10.3NLKK829 pKa = 9.25FQLNGRR835 pKa = 11.84YY836 pKa = 9.12LSQSGGSFTSVSSPADD852 pKa = 3.06ISAVLISGDD861 pKa = 3.7LGATNNCLLTTSLDD875 pKa = 3.34ATTFDD880 pKa = 5.04CSNLGDD886 pKa = 3.91NEE888 pKa = 4.18VTLTVTDD895 pKa = 4.41GSGNSSSATAIVTVIDD911 pKa = 4.51NIAPTIVLKK920 pKa = 10.96DD921 pKa = 3.33VTVEE925 pKa = 3.89LNAEE929 pKa = 4.14GTVSVEE935 pKa = 4.59ASAFDD940 pKa = 3.53NGSFDD945 pKa = 3.41NCGEE949 pKa = 4.31VTFSTNLVEE958 pKa = 5.04ILSCGALGLNEE969 pKa = 4.07VTVTVTDD976 pKa = 3.69ASGNQSQNTAILTVLDD992 pKa = 4.08NLSPAVVTQDD1002 pKa = 3.11FTVVLNANGEE1012 pKa = 4.21ATITAAQVDD1021 pKa = 4.2NGSADD1026 pKa = 3.33NCGVDD1031 pKa = 4.04TLALDD1036 pKa = 3.5ITTFDD1041 pKa = 4.27CSNLGEE1047 pKa = 4.23NTVTLTATDD1056 pKa = 4.1GSGKK1060 pKa = 9.72QATATAIVTVVDD1072 pKa = 3.65NSLPTAVAQNLTVEE1086 pKa = 4.18LDD1088 pKa = 3.53EE1089 pKa = 5.56FGAASITAAQVDD1101 pKa = 4.03NGSSDD1106 pKa = 3.33NCGVEE1111 pKa = 4.31TLALDD1116 pKa = 3.77VTSFDD1121 pKa = 4.49CSNLGEE1127 pKa = 4.3NTVTLRR1133 pKa = 11.84VTDD1136 pKa = 4.12GSGNQATSTAVITVIDD1152 pKa = 4.21ALAPALVTADD1162 pKa = 3.13ITVSLDD1168 pKa = 3.21EE1169 pKa = 4.52NATVSIEE1176 pKa = 3.91PGNLVDD1182 pKa = 4.64TLEE1185 pKa = 5.03DD1186 pKa = 3.52NCSLAGEE1193 pKa = 4.25ITLALDD1199 pKa = 3.17QDD1201 pKa = 4.2TFTATGIYY1209 pKa = 9.25VVNITATDD1217 pKa = 3.41ASGNTTTEE1225 pKa = 4.17SAEE1228 pKa = 4.08VTVDD1232 pKa = 3.2STLSIDD1238 pKa = 4.4QVDD1241 pKa = 3.46SDD1243 pKa = 4.5IQVKK1247 pKa = 10.01LYY1249 pKa = 9.6PNPTTDD1255 pKa = 3.02HH1256 pKa = 6.76LFFEE1260 pKa = 4.9VSNTEE1265 pKa = 4.45LIHH1268 pKa = 5.15ITIYY1272 pKa = 11.35DD1273 pKa = 3.72MNGKK1277 pKa = 8.77LVKK1280 pKa = 9.85TSQEE1284 pKa = 3.94SHH1286 pKa = 6.22SVDD1289 pKa = 3.36VTNLSSGVYY1298 pKa = 8.62FAKK1301 pKa = 10.51VDD1303 pKa = 3.66AKK1305 pKa = 11.0GGNTLKK1311 pKa = 10.57ILRR1314 pKa = 11.84FIKK1317 pKa = 10.44KK1318 pKa = 9.73
Molecular weight: 137.22 kDa
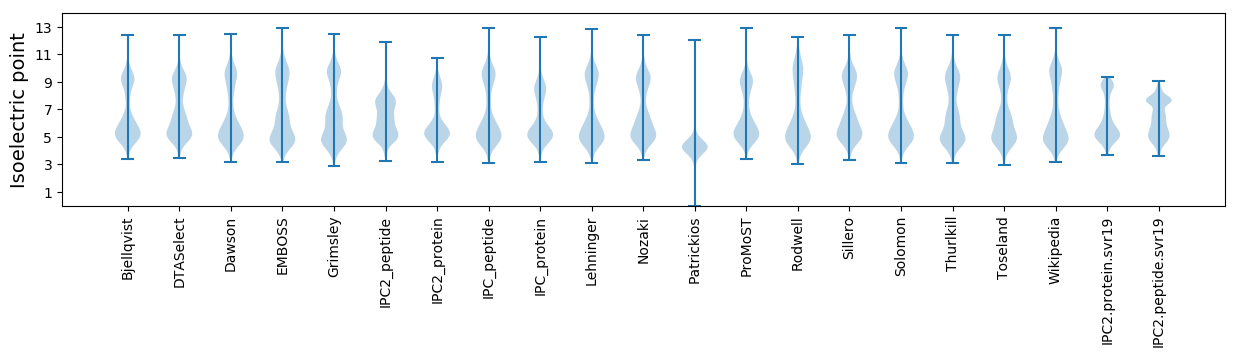
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7XSZ8|A0A1G7XSZ8_9FLAO Peptide methionine sulfoxide reductase MsrA OS=Psychroflexus sediminis OX=470826 GN=msrA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNKK10 pKa = 8.92KK11 pKa = 9.95RR12 pKa = 11.84KK13 pKa = 8.04NKK15 pKa = 9.86HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MSSVSGRR28 pKa = 11.84KK29 pKa = 7.69VLKK32 pKa = 10.02RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.37MSLRR51 pKa = 11.84KK52 pKa = 9.51RR53 pKa = 3.5
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNKK10 pKa = 8.92KK11 pKa = 9.95RR12 pKa = 11.84KK13 pKa = 8.04NKK15 pKa = 9.86HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MSSVSGRR28 pKa = 11.84KK29 pKa = 7.69VLKK32 pKa = 10.02RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.37MSLRR51 pKa = 11.84KK52 pKa = 9.51RR53 pKa = 3.5
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
886846 |
40 |
2412 |
334.0 |
37.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.252 ± 0.042 | 0.643 ± 0.013 |
5.76 ± 0.035 | 7.271 ± 0.043 |
5.414 ± 0.039 | 6.281 ± 0.044 |
1.851 ± 0.023 | 7.59 ± 0.047 |
7.56 ± 0.058 | 9.719 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.193 ± 0.023 | 5.458 ± 0.051 |
3.407 ± 0.027 | 3.639 ± 0.027 |
3.656 ± 0.031 | 6.959 ± 0.042 |
5.343 ± 0.039 | 6.086 ± 0.038 |
1.016 ± 0.017 | 3.901 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
