
Curtobacterium sp. MCBD17_040
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Curtobacterium; unclassified Curtobacterium
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
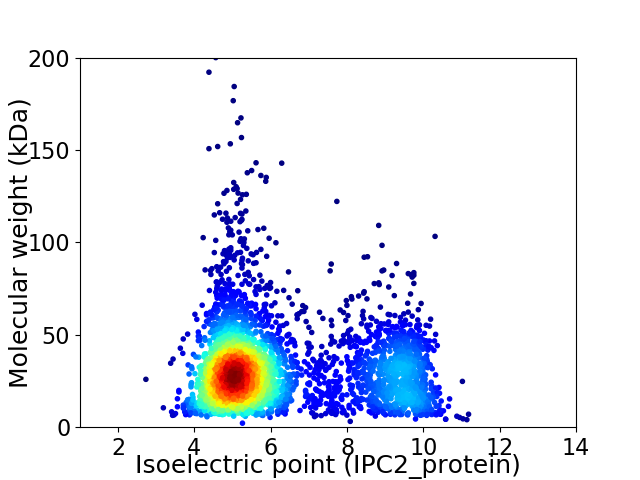
Virtual 2D-PAGE plot for 3754 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
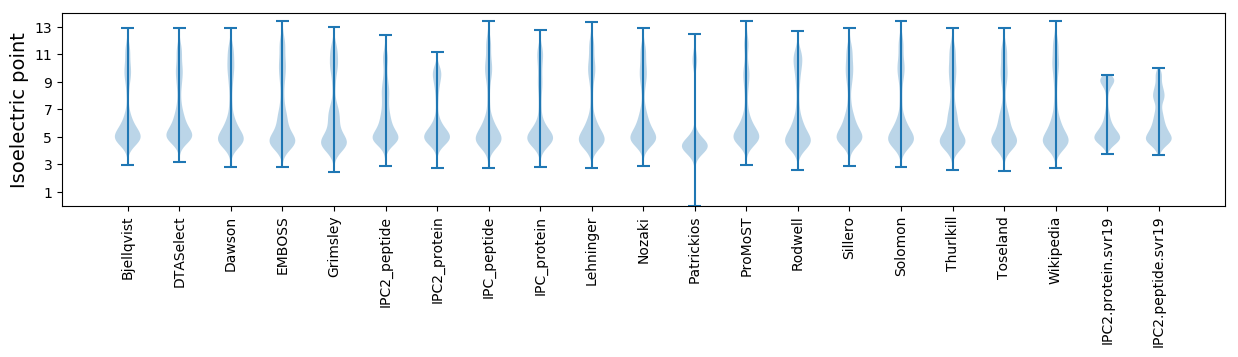
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W1ZRA1|A0A2W1ZRA1_9MICO Uncharacterized protein OS=Curtobacterium sp. MCBD17_040 OX=2175674 GN=DEI94_15800 PE=4 SV=1
MM1 pKa = 7.81AGTFSGIQSAYY12 pKa = 10.55SGLTAAQTALNVVGQNIDD30 pKa = 3.41NASTAGYY37 pKa = 8.94VRR39 pKa = 11.84QRR41 pKa = 11.84VEE43 pKa = 3.77QTAVGSPAQVGLAGGTTQAGQGVLVTAISNLSSVFLDD80 pKa = 3.54SQVRR84 pKa = 11.84AASAANSYY92 pKa = 9.36TGARR96 pKa = 11.84ATGYY100 pKa = 9.39TSVEE104 pKa = 4.16TALGEE109 pKa = 4.26PSDD112 pKa = 4.78DD113 pKa = 3.75ALSSQMNSFWSAWSDD128 pKa = 3.67VSNQPGNDD136 pKa = 3.49APVGALLGQAQTLTTQIGDD155 pKa = 4.16AYY157 pKa = 10.98RR158 pKa = 11.84SMQIGWSNTYY168 pKa = 10.1QSAGTEE174 pKa = 3.91VDD176 pKa = 3.24QLNALTSQVATLNGQIRR193 pKa = 11.84STIASGNNANEE204 pKa = 4.16LTDD207 pKa = 3.82EE208 pKa = 4.52RR209 pKa = 11.84SQLAAQISALAGGSVAQNSDD229 pKa = 2.78NTINITIGGSNVVDD243 pKa = 3.64GTNARR248 pKa = 11.84QVSLSGADD256 pKa = 3.41TLEE259 pKa = 4.26NASANPVTITWSDD272 pKa = 3.61QVGGAPLSLTGGSIAGNLSLLAGANPAGTGGAYY305 pKa = 9.9AQAADD310 pKa = 4.36ALNTLATSIATSVNAVSEE328 pKa = 4.43TGATANGTTGTDD340 pKa = 4.0FFSLTAGQPAALSISVIPTNADD362 pKa = 2.78GVAVGTPGAGANDD375 pKa = 3.92GSVADD380 pKa = 4.5QISQIGTGATSPSNQWSTYY399 pKa = 7.79VVSLGTQSSVEE410 pKa = 4.31TQQSTLATTTYY421 pKa = 10.49NSAVTSQQSQEE432 pKa = 4.26GVDD435 pKa = 3.85LDD437 pKa = 4.12EE438 pKa = 5.32EE439 pKa = 5.01SMNLLEE445 pKa = 5.13AQHH448 pKa = 6.98AYY450 pKa = 10.13QGAARR455 pKa = 11.84VMTAMDD461 pKa = 3.44QMLDD465 pKa = 3.21QLINHH470 pKa = 7.32TGTVGISS477 pKa = 2.98
MM1 pKa = 7.81AGTFSGIQSAYY12 pKa = 10.55SGLTAAQTALNVVGQNIDD30 pKa = 3.41NASTAGYY37 pKa = 8.94VRR39 pKa = 11.84QRR41 pKa = 11.84VEE43 pKa = 3.77QTAVGSPAQVGLAGGTTQAGQGVLVTAISNLSSVFLDD80 pKa = 3.54SQVRR84 pKa = 11.84AASAANSYY92 pKa = 9.36TGARR96 pKa = 11.84ATGYY100 pKa = 9.39TSVEE104 pKa = 4.16TALGEE109 pKa = 4.26PSDD112 pKa = 4.78DD113 pKa = 3.75ALSSQMNSFWSAWSDD128 pKa = 3.67VSNQPGNDD136 pKa = 3.49APVGALLGQAQTLTTQIGDD155 pKa = 4.16AYY157 pKa = 10.98RR158 pKa = 11.84SMQIGWSNTYY168 pKa = 10.1QSAGTEE174 pKa = 3.91VDD176 pKa = 3.24QLNALTSQVATLNGQIRR193 pKa = 11.84STIASGNNANEE204 pKa = 4.16LTDD207 pKa = 3.82EE208 pKa = 4.52RR209 pKa = 11.84SQLAAQISALAGGSVAQNSDD229 pKa = 2.78NTINITIGGSNVVDD243 pKa = 3.64GTNARR248 pKa = 11.84QVSLSGADD256 pKa = 3.41TLEE259 pKa = 4.26NASANPVTITWSDD272 pKa = 3.61QVGGAPLSLTGGSIAGNLSLLAGANPAGTGGAYY305 pKa = 9.9AQAADD310 pKa = 4.36ALNTLATSIATSVNAVSEE328 pKa = 4.43TGATANGTTGTDD340 pKa = 4.0FFSLTAGQPAALSISVIPTNADD362 pKa = 2.78GVAVGTPGAGANDD375 pKa = 3.92GSVADD380 pKa = 4.5QISQIGTGATSPSNQWSTYY399 pKa = 7.79VVSLGTQSSVEE410 pKa = 4.31TQQSTLATTTYY421 pKa = 10.49NSAVTSQQSQEE432 pKa = 4.26GVDD435 pKa = 3.85LDD437 pKa = 4.12EE438 pKa = 5.32EE439 pKa = 5.01SMNLLEE445 pKa = 5.13AQHH448 pKa = 6.98AYY450 pKa = 10.13QGAARR455 pKa = 11.84VMTAMDD461 pKa = 3.44QMLDD465 pKa = 3.21QLINHH470 pKa = 7.32TGTVGISS477 pKa = 2.98
Molecular weight: 47.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W1YIC5|A0A2W1YIC5_9MICO Uncharacterized protein OS=Curtobacterium sp. MCBD17_040 OX=2175674 GN=DEI94_07445 PE=4 SV=1
MM1 pKa = 7.8PRR3 pKa = 11.84RR4 pKa = 11.84CSTARR9 pKa = 11.84GGPGTPPTRR18 pKa = 11.84RR19 pKa = 11.84TPTARR24 pKa = 11.84TPRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 5.61RR33 pKa = 11.84AAPTRR38 pKa = 11.84VVATNVPRR46 pKa = 11.84RR47 pKa = 11.84APPPPGRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84AARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84AA63 pKa = 3.21
MM1 pKa = 7.8PRR3 pKa = 11.84RR4 pKa = 11.84CSTARR9 pKa = 11.84GGPGTPPTRR18 pKa = 11.84RR19 pKa = 11.84TPTARR24 pKa = 11.84TPRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 5.61RR33 pKa = 11.84AAPTRR38 pKa = 11.84VVATNVPRR46 pKa = 11.84RR47 pKa = 11.84APPPPGRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84AARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84AA63 pKa = 3.21
Molecular weight: 7.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1159833 |
19 |
1947 |
309.0 |
32.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.702 ± 0.061 | 0.489 ± 0.009 |
6.534 ± 0.037 | 4.966 ± 0.042 |
2.933 ± 0.025 | 9.008 ± 0.037 |
2.206 ± 0.022 | 3.785 ± 0.029 |
1.745 ± 0.031 | 9.374 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.694 ± 0.015 | 1.876 ± 0.025 |
5.568 ± 0.034 | 2.881 ± 0.027 |
7.775 ± 0.053 | 5.542 ± 0.042 |
6.779 ± 0.05 | 9.717 ± 0.047 |
1.506 ± 0.018 | 1.921 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
