
Nanohaloarchaea archaeon
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Nanohaloarchaea; unclassified Nanohaloarchaea
Average proteome isoelectric point is 5.15
Get precalculated fractions of proteins
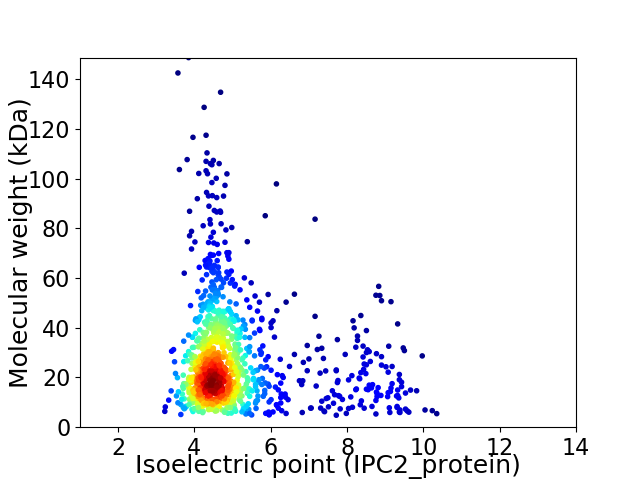
Virtual 2D-PAGE plot for 1074 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N0NRN8|A0A6N0NRN8_9EURY Glycosyltransferase OS=Nanohaloarchaea archaeon OX=2594798 GN=GKQ38_03465 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84KK3 pKa = 9.38YY4 pKa = 10.56LALLGTLVFLLLSIPAATASTVTWSDD30 pKa = 3.08SSDD33 pKa = 3.03WNNAVSDD40 pKa = 3.82FGVVHH45 pKa = 6.63EE46 pKa = 5.05SVTNTDD52 pKa = 3.58HH53 pKa = 7.42NDD55 pKa = 3.11AGKK58 pKa = 9.4LQKK61 pKa = 10.41GYY63 pKa = 9.5PIQNPVVSYY72 pKa = 10.94GLVGYY77 pKa = 9.07WPLQEE82 pKa = 5.62DD83 pKa = 4.22SGDD86 pKa = 3.48TAYY89 pKa = 10.81DD90 pKa = 3.55FSGYY94 pKa = 10.58NDD96 pKa = 3.89ASASAPTQGSSGILGSSSYY115 pKa = 11.26YY116 pKa = 10.74FNTEE120 pKa = 3.57SADD123 pKa = 3.17ASTIDD128 pKa = 3.78LEE130 pKa = 4.64YY131 pKa = 11.06YY132 pKa = 10.72SFNAWINDD140 pKa = 3.63EE141 pKa = 3.81GSTYY145 pKa = 9.61YY146 pKa = 10.82ARR148 pKa = 11.84AMSNGRR154 pKa = 11.84DD155 pKa = 3.7GTDD158 pKa = 3.27PSATYY163 pKa = 11.59AMGSRR168 pKa = 11.84NGWPTAVMRR177 pKa = 11.84KK178 pKa = 9.5NGNTYY183 pKa = 10.09TIGSGLNPSDD193 pKa = 2.86WTMVTVTYY201 pKa = 10.83DD202 pKa = 3.2GDD204 pKa = 3.89TLKK207 pKa = 11.07VYY209 pKa = 10.5TDD211 pKa = 3.78GSLTGSSTAPSGPIGDD227 pKa = 3.52VTGPFNLGSTEE238 pKa = 4.09RR239 pKa = 11.84YY240 pKa = 8.47PGNEE244 pKa = 4.22GITGYY249 pKa = 10.29LQEE252 pKa = 4.28ASLYY256 pKa = 10.24NRR258 pKa = 11.84ALSASEE264 pKa = 3.78IQSLYY269 pKa = 10.32EE270 pKa = 3.82VAATPGTLTTGWKK283 pKa = 8.37TFSSNRR289 pKa = 11.84DD290 pKa = 3.4MNSLQLEE297 pKa = 4.35NVQASLNGGSITVYY311 pKa = 10.81VEE313 pKa = 4.06TQNGEE318 pKa = 3.89VSDD321 pKa = 5.18PISLDD326 pKa = 3.39GSGGPYY332 pKa = 10.15SVTGLSTEE340 pKa = 4.0EE341 pKa = 3.74DD342 pKa = 3.46QFRR345 pKa = 11.84LDD347 pKa = 3.41IRR349 pKa = 11.84LDD351 pKa = 3.64NSDD354 pKa = 3.56VTSTPTLSAIDD365 pKa = 3.66LTGNKK370 pKa = 9.64ANSPPAAPSNEE381 pKa = 4.0DD382 pKa = 3.47PADD385 pKa = 3.72NADD388 pKa = 4.09PVSLSPSLSVFVDD401 pKa = 3.98DD402 pKa = 5.69PDD404 pKa = 5.59GDD406 pKa = 4.25SMDD409 pKa = 3.21VTFYY413 pKa = 11.67VDD415 pKa = 3.36TNNDD419 pKa = 2.78GSYY422 pKa = 8.26EE423 pKa = 4.21TTKK426 pKa = 10.71TNTGVASGSTTSVSLSSLATSQTVNWYY453 pKa = 9.64VVADD457 pKa = 4.44DD458 pKa = 5.33GRR460 pKa = 11.84ATTQSSTWSFTTVGVPSITSAPSPSDD486 pKa = 3.17GAVDD490 pKa = 3.57VSTSKK495 pKa = 11.03NLAIDD500 pKa = 3.52VSHH503 pKa = 7.89PDD505 pKa = 3.48NLDD508 pKa = 3.26MTVEE512 pKa = 4.44FSMDD516 pKa = 2.94GSYY519 pKa = 10.86VGSDD523 pKa = 3.21TVAGSGTASYY533 pKa = 10.66SPSLGYY539 pKa = 10.11SEE541 pKa = 4.59DD542 pKa = 3.96HH543 pKa = 6.29SWSVDD548 pKa = 3.19VYY550 pKa = 8.26TQGYY554 pKa = 6.77STQTSGGAWNFHH566 pKa = 5.13TVHH569 pKa = 6.84DD570 pKa = 4.74VSASLSGPSKK580 pKa = 10.76SPASIDD586 pKa = 3.44PTLSATVSHH595 pKa = 6.86SDD597 pKa = 3.04SAEE600 pKa = 3.94TVTVDD605 pKa = 5.55FVNTDD610 pKa = 3.12NGNTICSVSGIGNAEE625 pKa = 4.11TATCDD630 pKa = 3.23TSGSSFGSSPGTTYY644 pKa = 10.72NWEE647 pKa = 4.09VQLSGDD653 pKa = 4.14EE654 pKa = 4.78GSEE657 pKa = 4.06SWTSSQRR664 pKa = 11.84SFTTISNPNVNRR676 pKa = 11.84VEE678 pKa = 4.28PGSNSGVSTDD688 pKa = 3.3QNLVVDD694 pKa = 4.61VSHH697 pKa = 8.05PDD699 pKa = 3.3GKK701 pKa = 10.78NMNVEE706 pKa = 3.84FMIRR710 pKa = 11.84KK711 pKa = 9.4AGQTSFSTVDD721 pKa = 2.98TKK723 pKa = 11.16TDD725 pKa = 2.99VSGQVSANPSLSDD738 pKa = 3.15GTSYY742 pKa = 10.53EE743 pKa = 3.55WRR745 pKa = 11.84VDD747 pKa = 3.28VRR749 pKa = 11.84TVGYY753 pKa = 8.28STDD756 pKa = 3.33TMSSIWGFTTNYY768 pKa = 8.53VPSVSGITKK777 pKa = 9.46YY778 pKa = 10.56DD779 pKa = 3.27ASDD782 pKa = 2.87GHH784 pKa = 6.71AFRR787 pKa = 11.84NVSAIISDD795 pKa = 3.54QDD797 pKa = 3.49GDD799 pKa = 4.04DD800 pKa = 4.3TIVDD804 pKa = 3.73ANLTVDD810 pKa = 3.4NDD812 pKa = 4.1GGLEE816 pKa = 4.1TYY818 pKa = 10.49VEE820 pKa = 4.17YY821 pKa = 11.22DD822 pKa = 3.18EE823 pKa = 5.64VSIDD827 pKa = 3.31RR828 pKa = 11.84SYY830 pKa = 11.97GGDD833 pKa = 3.42DD834 pKa = 3.3NQANVTFGRR843 pKa = 11.84VNVGDD848 pKa = 4.44SNWGLTGIDD857 pKa = 3.94LQVEE861 pKa = 4.96GIDD864 pKa = 3.48QEE866 pKa = 4.93GNVGSNTLSNVQFPNHH882 pKa = 6.68APTVASGYY890 pKa = 9.91SYY892 pKa = 11.31SDD894 pKa = 3.24SPQAHH899 pKa = 7.15SYY901 pKa = 8.86TLSIDD906 pKa = 3.62AQDD909 pKa = 3.74VDD911 pKa = 4.32DD912 pKa = 4.55TDD914 pKa = 4.63EE915 pKa = 5.1EE916 pKa = 4.52IASCTVDD923 pKa = 4.69HH924 pKa = 7.26NDD926 pKa = 3.77GDD928 pKa = 4.49GNSYY932 pKa = 8.98TGKK935 pKa = 10.4AGSLASGEE943 pKa = 4.26TATCSFTIDD952 pKa = 3.66NSTSGYY958 pKa = 10.5RR959 pKa = 11.84FGEE962 pKa = 4.45MINHH966 pKa = 6.5RR967 pKa = 11.84VTFKK971 pKa = 10.55DD972 pKa = 3.22AHH974 pKa = 6.12GKK976 pKa = 7.58TVTSTWASHH985 pKa = 7.23RR986 pKa = 11.84IPNSLPSATNLRR998 pKa = 11.84PASDD1002 pKa = 3.61NTSYY1006 pKa = 11.44DD1007 pKa = 3.56ADD1009 pKa = 4.03LKK1011 pKa = 10.42ATYY1014 pKa = 9.99NDD1016 pKa = 3.81PDD1018 pKa = 4.77GDD1020 pKa = 3.95VGNLTFYY1027 pKa = 9.88NTTSGEE1033 pKa = 4.07EE1034 pKa = 3.92LGAVADD1040 pKa = 4.73LAPGEE1045 pKa = 4.3QGTVDD1050 pKa = 4.12LVGEE1054 pKa = 4.44LAPGTTYY1061 pKa = 11.34NFTVEE1066 pKa = 4.59ASDD1069 pKa = 4.35GVNSTNSTEE1078 pKa = 4.09NFTTIYY1084 pKa = 10.56RR1085 pKa = 11.84PDD1087 pKa = 3.47EE1088 pKa = 4.62PYY1090 pKa = 11.28DD1091 pKa = 3.78PFPEE1095 pKa = 4.57NDD1097 pKa = 3.5TVIDD1101 pKa = 3.63TTTRR1105 pKa = 11.84SGDD1108 pKa = 3.78DD1109 pKa = 2.76IAASVKK1115 pKa = 9.69VVQDD1119 pKa = 4.16DD1120 pKa = 4.05GHH1122 pKa = 5.71TMNVQFINASDD1133 pKa = 4.24DD1134 pKa = 3.91SPLGTDD1140 pKa = 3.57YY1141 pKa = 11.32DD1142 pKa = 4.22VEE1144 pKa = 4.65SGTRR1148 pKa = 11.84AKK1150 pKa = 9.56LTSIGNTLRR1159 pKa = 11.84DD1160 pKa = 3.93EE1161 pKa = 4.6TNTTYY1166 pKa = 10.71RR1167 pKa = 11.84WYY1169 pKa = 10.67AVATDD1174 pKa = 3.59QTTGEE1179 pKa = 4.45SVRR1182 pKa = 11.84SDD1184 pKa = 2.99TFVFRR1189 pKa = 11.84TPEE1192 pKa = 3.8VGEE1195 pKa = 3.94VRR1197 pKa = 11.84FDD1199 pKa = 3.55VMQGRR1204 pKa = 11.84NEE1206 pKa = 3.94NMDD1209 pKa = 2.87ITGNSDD1215 pKa = 2.44NGYY1218 pKa = 9.73RR1219 pKa = 11.84KK1220 pKa = 9.52IKK1222 pKa = 10.74FEE1224 pKa = 4.17VTSTEE1229 pKa = 3.78MDD1231 pKa = 5.2PIPTVIVNEE1240 pKa = 4.28TGTGNMIKK1248 pKa = 9.81SWSNVANNSVLTIDD1262 pKa = 4.69LVQDD1266 pKa = 4.74GSQDD1270 pKa = 3.19WNLDD1274 pKa = 3.16TDD1276 pKa = 4.68EE1277 pKa = 5.04KK1278 pKa = 11.48YY1279 pKa = 10.67EE1280 pKa = 4.07WTVEE1284 pKa = 3.88ARR1286 pKa = 11.84DD1287 pKa = 3.71GSNILGNRR1295 pKa = 11.84SYY1297 pKa = 11.24DD1298 pKa = 2.87IYY1300 pKa = 10.53TYY1302 pKa = 11.28NVTLDD1307 pKa = 2.89WDD1309 pKa = 4.01RR1310 pKa = 11.84AEE1312 pKa = 4.66KK1313 pKa = 10.54YY1314 pKa = 10.52YY1315 pKa = 11.19NVYY1318 pKa = 10.43QYY1320 pKa = 11.12DD1321 pKa = 3.58IYY1323 pKa = 10.84RR1324 pKa = 11.84AEE1326 pKa = 3.95QRR1328 pKa = 11.84SGVDD1332 pKa = 3.09LTFNYY1337 pKa = 10.57GNGDD1341 pKa = 3.77YY1342 pKa = 10.92NLVGSLPEE1350 pKa = 3.85TSFNDD1355 pKa = 3.46SGPGIVPDD1363 pKa = 3.89HH1364 pKa = 6.77TGTFCWKK1371 pKa = 9.76VAASNPTGSSPAIPSGEE1388 pKa = 4.33GNCKK1392 pKa = 8.47TLNN1395 pKa = 3.45
MM1 pKa = 7.76RR2 pKa = 11.84KK3 pKa = 9.38YY4 pKa = 10.56LALLGTLVFLLLSIPAATASTVTWSDD30 pKa = 3.08SSDD33 pKa = 3.03WNNAVSDD40 pKa = 3.82FGVVHH45 pKa = 6.63EE46 pKa = 5.05SVTNTDD52 pKa = 3.58HH53 pKa = 7.42NDD55 pKa = 3.11AGKK58 pKa = 9.4LQKK61 pKa = 10.41GYY63 pKa = 9.5PIQNPVVSYY72 pKa = 10.94GLVGYY77 pKa = 9.07WPLQEE82 pKa = 5.62DD83 pKa = 4.22SGDD86 pKa = 3.48TAYY89 pKa = 10.81DD90 pKa = 3.55FSGYY94 pKa = 10.58NDD96 pKa = 3.89ASASAPTQGSSGILGSSSYY115 pKa = 11.26YY116 pKa = 10.74FNTEE120 pKa = 3.57SADD123 pKa = 3.17ASTIDD128 pKa = 3.78LEE130 pKa = 4.64YY131 pKa = 11.06YY132 pKa = 10.72SFNAWINDD140 pKa = 3.63EE141 pKa = 3.81GSTYY145 pKa = 9.61YY146 pKa = 10.82ARR148 pKa = 11.84AMSNGRR154 pKa = 11.84DD155 pKa = 3.7GTDD158 pKa = 3.27PSATYY163 pKa = 11.59AMGSRR168 pKa = 11.84NGWPTAVMRR177 pKa = 11.84KK178 pKa = 9.5NGNTYY183 pKa = 10.09TIGSGLNPSDD193 pKa = 2.86WTMVTVTYY201 pKa = 10.83DD202 pKa = 3.2GDD204 pKa = 3.89TLKK207 pKa = 11.07VYY209 pKa = 10.5TDD211 pKa = 3.78GSLTGSSTAPSGPIGDD227 pKa = 3.52VTGPFNLGSTEE238 pKa = 4.09RR239 pKa = 11.84YY240 pKa = 8.47PGNEE244 pKa = 4.22GITGYY249 pKa = 10.29LQEE252 pKa = 4.28ASLYY256 pKa = 10.24NRR258 pKa = 11.84ALSASEE264 pKa = 3.78IQSLYY269 pKa = 10.32EE270 pKa = 3.82VAATPGTLTTGWKK283 pKa = 8.37TFSSNRR289 pKa = 11.84DD290 pKa = 3.4MNSLQLEE297 pKa = 4.35NVQASLNGGSITVYY311 pKa = 10.81VEE313 pKa = 4.06TQNGEE318 pKa = 3.89VSDD321 pKa = 5.18PISLDD326 pKa = 3.39GSGGPYY332 pKa = 10.15SVTGLSTEE340 pKa = 4.0EE341 pKa = 3.74DD342 pKa = 3.46QFRR345 pKa = 11.84LDD347 pKa = 3.41IRR349 pKa = 11.84LDD351 pKa = 3.64NSDD354 pKa = 3.56VTSTPTLSAIDD365 pKa = 3.66LTGNKK370 pKa = 9.64ANSPPAAPSNEE381 pKa = 4.0DD382 pKa = 3.47PADD385 pKa = 3.72NADD388 pKa = 4.09PVSLSPSLSVFVDD401 pKa = 3.98DD402 pKa = 5.69PDD404 pKa = 5.59GDD406 pKa = 4.25SMDD409 pKa = 3.21VTFYY413 pKa = 11.67VDD415 pKa = 3.36TNNDD419 pKa = 2.78GSYY422 pKa = 8.26EE423 pKa = 4.21TTKK426 pKa = 10.71TNTGVASGSTTSVSLSSLATSQTVNWYY453 pKa = 9.64VVADD457 pKa = 4.44DD458 pKa = 5.33GRR460 pKa = 11.84ATTQSSTWSFTTVGVPSITSAPSPSDD486 pKa = 3.17GAVDD490 pKa = 3.57VSTSKK495 pKa = 11.03NLAIDD500 pKa = 3.52VSHH503 pKa = 7.89PDD505 pKa = 3.48NLDD508 pKa = 3.26MTVEE512 pKa = 4.44FSMDD516 pKa = 2.94GSYY519 pKa = 10.86VGSDD523 pKa = 3.21TVAGSGTASYY533 pKa = 10.66SPSLGYY539 pKa = 10.11SEE541 pKa = 4.59DD542 pKa = 3.96HH543 pKa = 6.29SWSVDD548 pKa = 3.19VYY550 pKa = 8.26TQGYY554 pKa = 6.77STQTSGGAWNFHH566 pKa = 5.13TVHH569 pKa = 6.84DD570 pKa = 4.74VSASLSGPSKK580 pKa = 10.76SPASIDD586 pKa = 3.44PTLSATVSHH595 pKa = 6.86SDD597 pKa = 3.04SAEE600 pKa = 3.94TVTVDD605 pKa = 5.55FVNTDD610 pKa = 3.12NGNTICSVSGIGNAEE625 pKa = 4.11TATCDD630 pKa = 3.23TSGSSFGSSPGTTYY644 pKa = 10.72NWEE647 pKa = 4.09VQLSGDD653 pKa = 4.14EE654 pKa = 4.78GSEE657 pKa = 4.06SWTSSQRR664 pKa = 11.84SFTTISNPNVNRR676 pKa = 11.84VEE678 pKa = 4.28PGSNSGVSTDD688 pKa = 3.3QNLVVDD694 pKa = 4.61VSHH697 pKa = 8.05PDD699 pKa = 3.3GKK701 pKa = 10.78NMNVEE706 pKa = 3.84FMIRR710 pKa = 11.84KK711 pKa = 9.4AGQTSFSTVDD721 pKa = 2.98TKK723 pKa = 11.16TDD725 pKa = 2.99VSGQVSANPSLSDD738 pKa = 3.15GTSYY742 pKa = 10.53EE743 pKa = 3.55WRR745 pKa = 11.84VDD747 pKa = 3.28VRR749 pKa = 11.84TVGYY753 pKa = 8.28STDD756 pKa = 3.33TMSSIWGFTTNYY768 pKa = 8.53VPSVSGITKK777 pKa = 9.46YY778 pKa = 10.56DD779 pKa = 3.27ASDD782 pKa = 2.87GHH784 pKa = 6.71AFRR787 pKa = 11.84NVSAIISDD795 pKa = 3.54QDD797 pKa = 3.49GDD799 pKa = 4.04DD800 pKa = 4.3TIVDD804 pKa = 3.73ANLTVDD810 pKa = 3.4NDD812 pKa = 4.1GGLEE816 pKa = 4.1TYY818 pKa = 10.49VEE820 pKa = 4.17YY821 pKa = 11.22DD822 pKa = 3.18EE823 pKa = 5.64VSIDD827 pKa = 3.31RR828 pKa = 11.84SYY830 pKa = 11.97GGDD833 pKa = 3.42DD834 pKa = 3.3NQANVTFGRR843 pKa = 11.84VNVGDD848 pKa = 4.44SNWGLTGIDD857 pKa = 3.94LQVEE861 pKa = 4.96GIDD864 pKa = 3.48QEE866 pKa = 4.93GNVGSNTLSNVQFPNHH882 pKa = 6.68APTVASGYY890 pKa = 9.91SYY892 pKa = 11.31SDD894 pKa = 3.24SPQAHH899 pKa = 7.15SYY901 pKa = 8.86TLSIDD906 pKa = 3.62AQDD909 pKa = 3.74VDD911 pKa = 4.32DD912 pKa = 4.55TDD914 pKa = 4.63EE915 pKa = 5.1EE916 pKa = 4.52IASCTVDD923 pKa = 4.69HH924 pKa = 7.26NDD926 pKa = 3.77GDD928 pKa = 4.49GNSYY932 pKa = 8.98TGKK935 pKa = 10.4AGSLASGEE943 pKa = 4.26TATCSFTIDD952 pKa = 3.66NSTSGYY958 pKa = 10.5RR959 pKa = 11.84FGEE962 pKa = 4.45MINHH966 pKa = 6.5RR967 pKa = 11.84VTFKK971 pKa = 10.55DD972 pKa = 3.22AHH974 pKa = 6.12GKK976 pKa = 7.58TVTSTWASHH985 pKa = 7.23RR986 pKa = 11.84IPNSLPSATNLRR998 pKa = 11.84PASDD1002 pKa = 3.61NTSYY1006 pKa = 11.44DD1007 pKa = 3.56ADD1009 pKa = 4.03LKK1011 pKa = 10.42ATYY1014 pKa = 9.99NDD1016 pKa = 3.81PDD1018 pKa = 4.77GDD1020 pKa = 3.95VGNLTFYY1027 pKa = 9.88NTTSGEE1033 pKa = 4.07EE1034 pKa = 3.92LGAVADD1040 pKa = 4.73LAPGEE1045 pKa = 4.3QGTVDD1050 pKa = 4.12LVGEE1054 pKa = 4.44LAPGTTYY1061 pKa = 11.34NFTVEE1066 pKa = 4.59ASDD1069 pKa = 4.35GVNSTNSTEE1078 pKa = 4.09NFTTIYY1084 pKa = 10.56RR1085 pKa = 11.84PDD1087 pKa = 3.47EE1088 pKa = 4.62PYY1090 pKa = 11.28DD1091 pKa = 3.78PFPEE1095 pKa = 4.57NDD1097 pKa = 3.5TVIDD1101 pKa = 3.63TTTRR1105 pKa = 11.84SGDD1108 pKa = 3.78DD1109 pKa = 2.76IAASVKK1115 pKa = 9.69VVQDD1119 pKa = 4.16DD1120 pKa = 4.05GHH1122 pKa = 5.71TMNVQFINASDD1133 pKa = 4.24DD1134 pKa = 3.91SPLGTDD1140 pKa = 3.57YY1141 pKa = 11.32DD1142 pKa = 4.22VEE1144 pKa = 4.65SGTRR1148 pKa = 11.84AKK1150 pKa = 9.56LTSIGNTLRR1159 pKa = 11.84DD1160 pKa = 3.93EE1161 pKa = 4.6TNTTYY1166 pKa = 10.71RR1167 pKa = 11.84WYY1169 pKa = 10.67AVATDD1174 pKa = 3.59QTTGEE1179 pKa = 4.45SVRR1182 pKa = 11.84SDD1184 pKa = 2.99TFVFRR1189 pKa = 11.84TPEE1192 pKa = 3.8VGEE1195 pKa = 3.94VRR1197 pKa = 11.84FDD1199 pKa = 3.55VMQGRR1204 pKa = 11.84NEE1206 pKa = 3.94NMDD1209 pKa = 2.87ITGNSDD1215 pKa = 2.44NGYY1218 pKa = 9.73RR1219 pKa = 11.84KK1220 pKa = 9.52IKK1222 pKa = 10.74FEE1224 pKa = 4.17VTSTEE1229 pKa = 3.78MDD1231 pKa = 5.2PIPTVIVNEE1240 pKa = 4.28TGTGNMIKK1248 pKa = 9.81SWSNVANNSVLTIDD1262 pKa = 4.69LVQDD1266 pKa = 4.74GSQDD1270 pKa = 3.19WNLDD1274 pKa = 3.16TDD1276 pKa = 4.68EE1277 pKa = 5.04KK1278 pKa = 11.48YY1279 pKa = 10.67EE1280 pKa = 4.07WTVEE1284 pKa = 3.88ARR1286 pKa = 11.84DD1287 pKa = 3.71GSNILGNRR1295 pKa = 11.84SYY1297 pKa = 11.24DD1298 pKa = 2.87IYY1300 pKa = 10.53TYY1302 pKa = 11.28NVTLDD1307 pKa = 2.89WDD1309 pKa = 4.01RR1310 pKa = 11.84AEE1312 pKa = 4.66KK1313 pKa = 10.54YY1314 pKa = 10.52YY1315 pKa = 11.19NVYY1318 pKa = 10.43QYY1320 pKa = 11.12DD1321 pKa = 3.58IYY1323 pKa = 10.84RR1324 pKa = 11.84AEE1326 pKa = 3.95QRR1328 pKa = 11.84SGVDD1332 pKa = 3.09LTFNYY1337 pKa = 10.57GNGDD1341 pKa = 3.77YY1342 pKa = 10.92NLVGSLPEE1350 pKa = 3.85TSFNDD1355 pKa = 3.46SGPGIVPDD1363 pKa = 3.89HH1364 pKa = 6.77TGTFCWKK1371 pKa = 9.76VAASNPTGSSPAIPSGEE1388 pKa = 4.33GNCKK1392 pKa = 8.47TLNN1395 pKa = 3.45
Molecular weight: 148.66 kDa
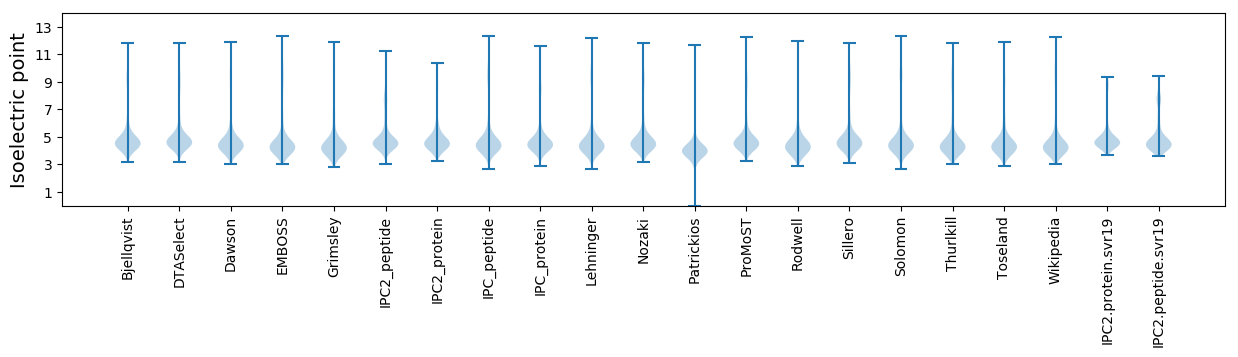
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N0NTP7|A0A6N0NTP7_9EURY TetR family transcriptional regulator OS=Nanohaloarchaea archaeon OX=2594798 GN=GKQ38_03365 PE=4 SV=1
MM1 pKa = 7.8LGILLLLMIAVPFTDD16 pKa = 5.9LYY18 pKa = 10.89ILLQIAGMIGFWQTLAIVIATGVIGAAIVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 3.8GRR52 pKa = 11.84HH53 pKa = 4.42VLKK56 pKa = 10.65KK57 pKa = 10.01LQRR60 pKa = 11.84SVTAGEE66 pKa = 4.07VSRR69 pKa = 11.84NVAEE73 pKa = 4.51GALLVIAGLLLITPGIITDD92 pKa = 3.43VTGFLLTFRR101 pKa = 11.84PLRR104 pKa = 11.84EE105 pKa = 3.61RR106 pKa = 11.84LVARR110 pKa = 11.84WISGNSGNIEE120 pKa = 3.74VEE122 pKa = 4.11VFRR125 pKa = 11.84FF126 pKa = 3.43
MM1 pKa = 7.8LGILLLLMIAVPFTDD16 pKa = 5.9LYY18 pKa = 10.89ILLQIAGMIGFWQTLAIVIATGVIGAAIVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 3.8GRR52 pKa = 11.84HH53 pKa = 4.42VLKK56 pKa = 10.65KK57 pKa = 10.01LQRR60 pKa = 11.84SVTAGEE66 pKa = 4.07VSRR69 pKa = 11.84NVAEE73 pKa = 4.51GALLVIAGLLLITPGIITDD92 pKa = 3.43VTGFLLTFRR101 pKa = 11.84PLRR104 pKa = 11.84EE105 pKa = 3.61RR106 pKa = 11.84LVARR110 pKa = 11.84WISGNSGNIEE120 pKa = 3.74VEE122 pKa = 4.11VFRR125 pKa = 11.84FF126 pKa = 3.43
Molecular weight: 13.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
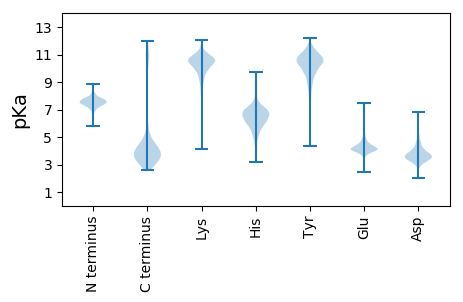
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
277001 |
45 |
1395 |
257.9 |
28.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.619 ± 0.075 | 0.608 ± 0.023 |
7.122 ± 0.079 | 11.233 ± 0.175 |
3.928 ± 0.061 | 7.204 ± 0.079 |
1.676 ± 0.038 | 6.087 ± 0.064 |
5.834 ± 0.081 | 8.324 ± 0.088 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.469 ± 0.039 | 4.107 ± 0.066 |
3.404 ± 0.041 | 3.491 ± 0.062 |
4.672 ± 0.059 | 6.838 ± 0.11 |
5.089 ± 0.09 | 7.062 ± 0.055 |
0.916 ± 0.028 | 3.319 ± 0.047 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
