
Croceibacter atlanticus (strain ATCC BAA-628 / HTCC2559 / KCTC 12090)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Croceibacter; Croceibacter atlanticus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
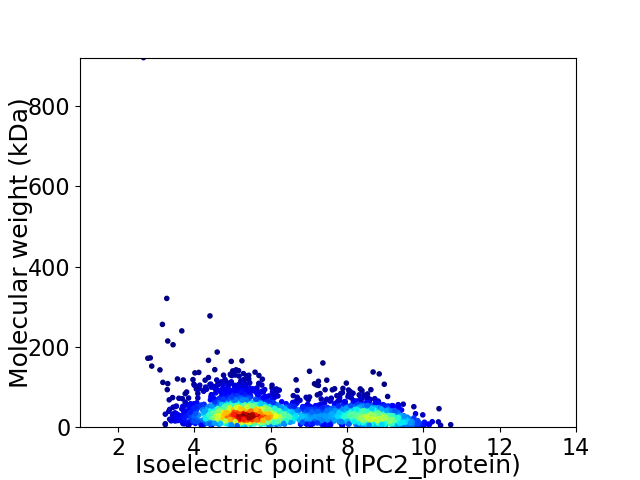
Virtual 2D-PAGE plot for 2702 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
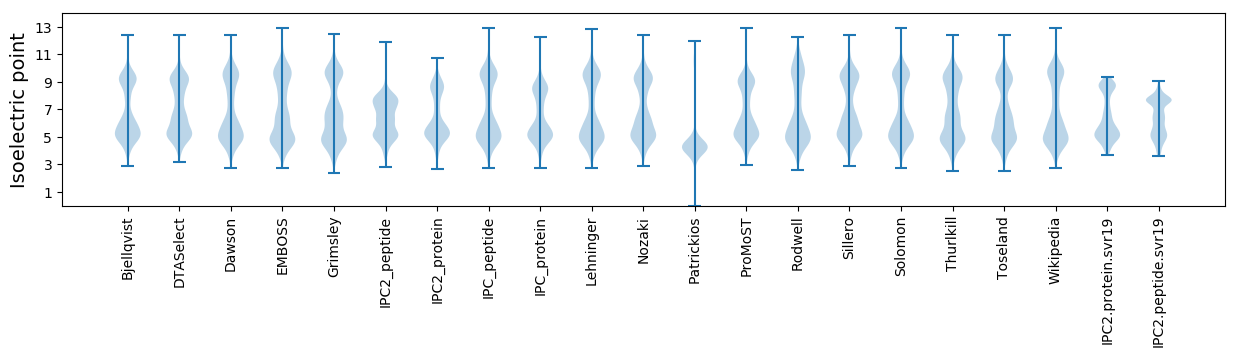
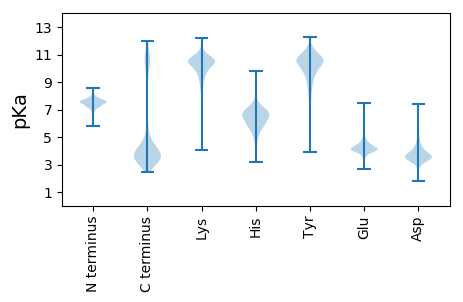
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3U7L0|A3U7L0_CROAH 30S ribosomal protein S12 OS=Croceibacter atlanticus (strain ATCC BAA-628 / HTCC2559 / KCTC 12090) OX=216432 GN=CA2559_05690 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.34KK3 pKa = 10.14IIYY6 pKa = 10.18LFTLILAVGTFQSCEE21 pKa = 3.94DD22 pKa = 3.83YY23 pKa = 11.6LDD25 pKa = 4.39INDD28 pKa = 4.98PLNDD32 pKa = 4.96PIEE35 pKa = 4.56SQVTPDD41 pKa = 5.01LILAGAMTGSFPSQSNNMNRR61 pKa = 11.84LGNVMMNNWAGNINAFTGGFNEE83 pKa = 4.63EE84 pKa = 3.95YY85 pKa = 10.58QLIITSTFYY94 pKa = 11.32NFIWDD99 pKa = 4.85GIYY102 pKa = 10.86LNVANFQAMIDD113 pKa = 3.56ADD115 pKa = 3.79FEE117 pKa = 4.42NYY119 pKa = 9.02EE120 pKa = 3.97NHH122 pKa = 6.64KK123 pKa = 10.82AIAKK127 pKa = 9.54IMKK130 pKa = 9.04SYY132 pKa = 10.02YY133 pKa = 7.49MQYY136 pKa = 10.88VVDD139 pKa = 5.42LYY141 pKa = 11.71GDD143 pKa = 3.67APYY146 pKa = 10.53TEE148 pKa = 5.15AFQGGDD154 pKa = 3.84NPTPTYY160 pKa = 10.93DD161 pKa = 4.61DD162 pKa = 3.84DD163 pKa = 4.14MEE165 pKa = 4.96IYY167 pKa = 10.1RR168 pKa = 11.84ALITEE173 pKa = 3.9VDD175 pKa = 3.54EE176 pKa = 5.09ALALIDD182 pKa = 4.89NNTDD186 pKa = 2.88LTNPVGSEE194 pKa = 3.64DD195 pKa = 3.53VVYY198 pKa = 10.8NGNMTNWVTFANTLKK213 pKa = 10.73LRR215 pKa = 11.84ILMRR219 pKa = 11.84QSTLAEE225 pKa = 4.2TDD227 pKa = 3.67GEE229 pKa = 4.58TASYY233 pKa = 10.7LATQFQALQDD243 pKa = 3.5ASFITTTAAINPGYY257 pKa = 11.13ANDD260 pKa = 4.05TGRR263 pKa = 11.84QNPFYY268 pKa = 9.94ATYY271 pKa = 10.36GFNVDD276 pKa = 4.05GSQTTSNRR284 pKa = 11.84FIVASLYY291 pKa = 10.85AEE293 pKa = 4.22QYY295 pKa = 11.4LDD297 pKa = 3.55GTLTGILDD305 pKa = 3.48PRR307 pKa = 11.84INEE310 pKa = 4.42LYY312 pKa = 9.88TPVDD316 pKa = 3.63GEE318 pKa = 4.5VQGVQQGVDD327 pKa = 3.58SNDD330 pKa = 3.06PSVPDD335 pKa = 4.2EE336 pKa = 4.25ISPLGEE342 pKa = 4.12GLLVDD347 pKa = 4.55ASQDD351 pKa = 3.27GVLFSAAEE359 pKa = 4.17SFFLQSEE366 pKa = 4.51AVFRR370 pKa = 11.84GYY372 pKa = 10.14IAGDD376 pKa = 3.34AKK378 pKa = 11.25GLFQDD383 pKa = 5.5GIRR386 pKa = 11.84SSFDD390 pKa = 2.96QLGIGAQAEE399 pKa = 4.38AYY401 pKa = 7.1ITNSDD406 pKa = 3.59GVDD409 pKa = 3.43EE410 pKa = 4.95IGWDD414 pKa = 3.56GSANKK419 pKa = 9.76IEE421 pKa = 5.77AIMTQKK427 pKa = 10.27WIATNGINAIEE438 pKa = 4.46SFIDD442 pKa = 3.34MNRR445 pKa = 11.84TNFPEE450 pKa = 4.07VPLAINAQRR459 pKa = 11.84DD460 pKa = 3.93KK461 pKa = 11.4KK462 pKa = 9.18PTRR465 pKa = 11.84LLYY468 pKa = 10.02PSSEE472 pKa = 4.54SIANAANKK480 pKa = 8.24PAQEE484 pKa = 4.04TDD486 pKa = 3.39DD487 pKa = 4.5AFNTYY492 pKa = 9.99IFWDD496 pKa = 3.68STQNN500 pKa = 3.24
MM1 pKa = 7.49KK2 pKa = 10.34KK3 pKa = 10.14IIYY6 pKa = 10.18LFTLILAVGTFQSCEE21 pKa = 3.94DD22 pKa = 3.83YY23 pKa = 11.6LDD25 pKa = 4.39INDD28 pKa = 4.98PLNDD32 pKa = 4.96PIEE35 pKa = 4.56SQVTPDD41 pKa = 5.01LILAGAMTGSFPSQSNNMNRR61 pKa = 11.84LGNVMMNNWAGNINAFTGGFNEE83 pKa = 4.63EE84 pKa = 3.95YY85 pKa = 10.58QLIITSTFYY94 pKa = 11.32NFIWDD99 pKa = 4.85GIYY102 pKa = 10.86LNVANFQAMIDD113 pKa = 3.56ADD115 pKa = 3.79FEE117 pKa = 4.42NYY119 pKa = 9.02EE120 pKa = 3.97NHH122 pKa = 6.64KK123 pKa = 10.82AIAKK127 pKa = 9.54IMKK130 pKa = 9.04SYY132 pKa = 10.02YY133 pKa = 7.49MQYY136 pKa = 10.88VVDD139 pKa = 5.42LYY141 pKa = 11.71GDD143 pKa = 3.67APYY146 pKa = 10.53TEE148 pKa = 5.15AFQGGDD154 pKa = 3.84NPTPTYY160 pKa = 10.93DD161 pKa = 4.61DD162 pKa = 3.84DD163 pKa = 4.14MEE165 pKa = 4.96IYY167 pKa = 10.1RR168 pKa = 11.84ALITEE173 pKa = 3.9VDD175 pKa = 3.54EE176 pKa = 5.09ALALIDD182 pKa = 4.89NNTDD186 pKa = 2.88LTNPVGSEE194 pKa = 3.64DD195 pKa = 3.53VVYY198 pKa = 10.8NGNMTNWVTFANTLKK213 pKa = 10.73LRR215 pKa = 11.84ILMRR219 pKa = 11.84QSTLAEE225 pKa = 4.2TDD227 pKa = 3.67GEE229 pKa = 4.58TASYY233 pKa = 10.7LATQFQALQDD243 pKa = 3.5ASFITTTAAINPGYY257 pKa = 11.13ANDD260 pKa = 4.05TGRR263 pKa = 11.84QNPFYY268 pKa = 9.94ATYY271 pKa = 10.36GFNVDD276 pKa = 4.05GSQTTSNRR284 pKa = 11.84FIVASLYY291 pKa = 10.85AEE293 pKa = 4.22QYY295 pKa = 11.4LDD297 pKa = 3.55GTLTGILDD305 pKa = 3.48PRR307 pKa = 11.84INEE310 pKa = 4.42LYY312 pKa = 9.88TPVDD316 pKa = 3.63GEE318 pKa = 4.5VQGVQQGVDD327 pKa = 3.58SNDD330 pKa = 3.06PSVPDD335 pKa = 4.2EE336 pKa = 4.25ISPLGEE342 pKa = 4.12GLLVDD347 pKa = 4.55ASQDD351 pKa = 3.27GVLFSAAEE359 pKa = 4.17SFFLQSEE366 pKa = 4.51AVFRR370 pKa = 11.84GYY372 pKa = 10.14IAGDD376 pKa = 3.34AKK378 pKa = 11.25GLFQDD383 pKa = 5.5GIRR386 pKa = 11.84SSFDD390 pKa = 2.96QLGIGAQAEE399 pKa = 4.38AYY401 pKa = 7.1ITNSDD406 pKa = 3.59GVDD409 pKa = 3.43EE410 pKa = 4.95IGWDD414 pKa = 3.56GSANKK419 pKa = 9.76IEE421 pKa = 5.77AIMTQKK427 pKa = 10.27WIATNGINAIEE438 pKa = 4.46SFIDD442 pKa = 3.34MNRR445 pKa = 11.84TNFPEE450 pKa = 4.07VPLAINAQRR459 pKa = 11.84DD460 pKa = 3.93KK461 pKa = 11.4KK462 pKa = 9.18PTRR465 pKa = 11.84LLYY468 pKa = 10.02PSSEE472 pKa = 4.54SIANAANKK480 pKa = 8.24PAQEE484 pKa = 4.04TDD486 pKa = 3.39DD487 pKa = 4.5AFNTYY492 pKa = 9.99IFWDD496 pKa = 3.68STQNN500 pKa = 3.24
Molecular weight: 55.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3UAT9|A3UAT9_CROAH Uncharacterized protein OS=Croceibacter atlanticus (strain ATCC BAA-628 / HTCC2559 / KCTC 12090) OX=216432 GN=CA2559_12833 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.83MRR49 pKa = 11.84HH50 pKa = 4.65KK51 pKa = 10.4HH52 pKa = 4.85
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.83MRR49 pKa = 11.84HH50 pKa = 4.65KK51 pKa = 10.4HH52 pKa = 4.85
Molecular weight: 6.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
907128 |
20 |
8918 |
335.7 |
37.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.491 ± 0.042 | 0.725 ± 0.017 |
5.93 ± 0.094 | 6.615 ± 0.046 |
5.045 ± 0.05 | 6.362 ± 0.067 |
1.727 ± 0.025 | 7.63 ± 0.047 |
7.315 ± 0.094 | 9.284 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.079 ± 0.028 | 6.264 ± 0.052 |
3.365 ± 0.032 | 3.469 ± 0.03 |
3.355 ± 0.047 | 6.565 ± 0.048 |
6.446 ± 0.111 | 6.427 ± 0.05 |
0.954 ± 0.016 | 3.952 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
