
Bovine faeces associated smacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Bonzesmacovirus; Bonzesmacovirus bovas1
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
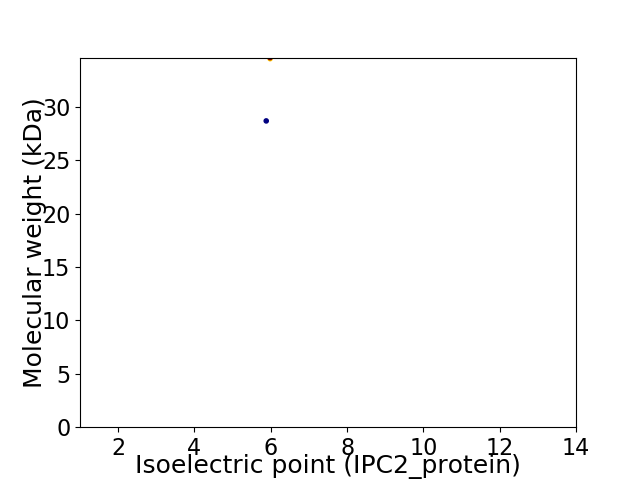
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
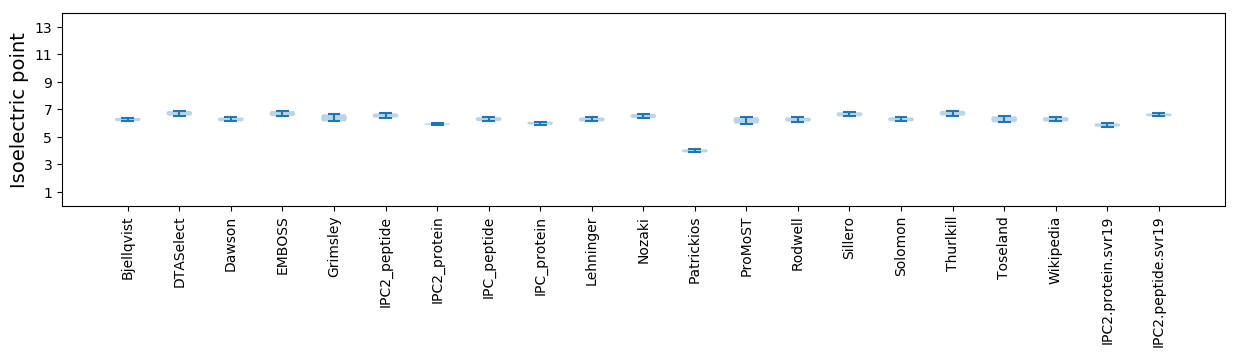
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HX16|A0A160HX16_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 1 OX=1843749 PE=4 SV=1
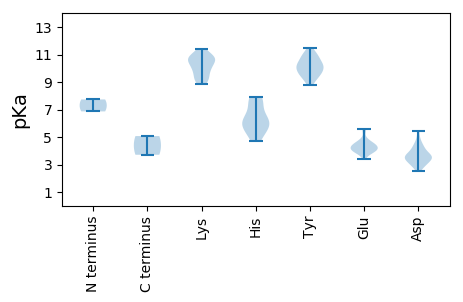
MM1 pKa = 7.73GFITEE6 pKa = 4.42DD7 pKa = 3.08AFFPEE12 pKa = 4.66FLEE15 pKa = 4.3KK16 pKa = 9.93TLQEE20 pKa = 4.46ALDD23 pKa = 4.54KK24 pKa = 10.92ICEE27 pKa = 3.98RR28 pKa = 11.84YY29 pKa = 10.13AYY31 pKa = 9.98GNEE34 pKa = 3.99VGKK37 pKa = 10.31DD38 pKa = 3.52GYY40 pKa = 10.23EE41 pKa = 3.89HH42 pKa = 5.92FQCRR46 pKa = 11.84IVCSKK51 pKa = 9.36PTDD54 pKa = 3.4EE55 pKa = 4.04RR56 pKa = 11.84ALRR59 pKa = 11.84VFLLSNGIPAHH70 pKa = 5.88TSPTQVRR77 pKa = 11.84NFEE80 pKa = 4.27YY81 pKa = 10.56VQKK84 pKa = 10.64EE85 pKa = 4.03GNLYY89 pKa = 10.38CSWEE93 pKa = 4.04AVLKK97 pKa = 9.03QFPRR101 pKa = 11.84GTPYY105 pKa = 9.25MWQLIAFSMWKK116 pKa = 9.32KK117 pKa = 10.5QNDD120 pKa = 3.61RR121 pKa = 11.84EE122 pKa = 4.25ILVITDD128 pKa = 2.97DD129 pKa = 3.75KK130 pKa = 11.4GRR132 pKa = 11.84HH133 pKa = 4.7GKK135 pKa = 8.87SWLRR139 pKa = 11.84KK140 pKa = 9.26YY141 pKa = 9.72MVACHH146 pKa = 6.36MGTFIPPLEE155 pKa = 4.17KK156 pKa = 10.98AEE158 pKa = 5.48DD159 pKa = 3.43IMACAMAKK167 pKa = 9.77PSKK170 pKa = 10.61GYY172 pKa = 10.2IIDD175 pKa = 3.59MPRR178 pKa = 11.84ADD180 pKa = 3.51GKK182 pKa = 10.0VRR184 pKa = 11.84KK185 pKa = 9.78AMWSAIEE192 pKa = 3.97QMKK195 pKa = 10.65DD196 pKa = 2.53GYY198 pKa = 11.02LYY200 pKa = 10.68DD201 pKa = 5.43KK202 pKa = 10.53RR203 pKa = 11.84YY204 pKa = 9.62SWQEE208 pKa = 3.41KK209 pKa = 8.89WIEE212 pKa = 3.85PPKK215 pKa = 10.91IMVFCNDD222 pKa = 4.26FDD224 pKa = 4.92PLMLSTDD231 pKa = 2.89RR232 pKa = 11.84WQSFDD237 pKa = 3.13ITDD240 pKa = 4.58FPQEE244 pKa = 3.71
MM1 pKa = 7.73GFITEE6 pKa = 4.42DD7 pKa = 3.08AFFPEE12 pKa = 4.66FLEE15 pKa = 4.3KK16 pKa = 9.93TLQEE20 pKa = 4.46ALDD23 pKa = 4.54KK24 pKa = 10.92ICEE27 pKa = 3.98RR28 pKa = 11.84YY29 pKa = 10.13AYY31 pKa = 9.98GNEE34 pKa = 3.99VGKK37 pKa = 10.31DD38 pKa = 3.52GYY40 pKa = 10.23EE41 pKa = 3.89HH42 pKa = 5.92FQCRR46 pKa = 11.84IVCSKK51 pKa = 9.36PTDD54 pKa = 3.4EE55 pKa = 4.04RR56 pKa = 11.84ALRR59 pKa = 11.84VFLLSNGIPAHH70 pKa = 5.88TSPTQVRR77 pKa = 11.84NFEE80 pKa = 4.27YY81 pKa = 10.56VQKK84 pKa = 10.64EE85 pKa = 4.03GNLYY89 pKa = 10.38CSWEE93 pKa = 4.04AVLKK97 pKa = 9.03QFPRR101 pKa = 11.84GTPYY105 pKa = 9.25MWQLIAFSMWKK116 pKa = 9.32KK117 pKa = 10.5QNDD120 pKa = 3.61RR121 pKa = 11.84EE122 pKa = 4.25ILVITDD128 pKa = 2.97DD129 pKa = 3.75KK130 pKa = 11.4GRR132 pKa = 11.84HH133 pKa = 4.7GKK135 pKa = 8.87SWLRR139 pKa = 11.84KK140 pKa = 9.26YY141 pKa = 9.72MVACHH146 pKa = 6.36MGTFIPPLEE155 pKa = 4.17KK156 pKa = 10.98AEE158 pKa = 5.48DD159 pKa = 3.43IMACAMAKK167 pKa = 9.77PSKK170 pKa = 10.61GYY172 pKa = 10.2IIDD175 pKa = 3.59MPRR178 pKa = 11.84ADD180 pKa = 3.51GKK182 pKa = 10.0VRR184 pKa = 11.84KK185 pKa = 9.78AMWSAIEE192 pKa = 3.97QMKK195 pKa = 10.65DD196 pKa = 2.53GYY198 pKa = 11.02LYY200 pKa = 10.68DD201 pKa = 5.43KK202 pKa = 10.53RR203 pKa = 11.84YY204 pKa = 9.62SWQEE208 pKa = 3.41KK209 pKa = 8.89WIEE212 pKa = 3.85PPKK215 pKa = 10.91IMVFCNDD222 pKa = 4.26FDD224 pKa = 4.92PLMLSTDD231 pKa = 2.89RR232 pKa = 11.84WQSFDD237 pKa = 3.13ITDD240 pKa = 4.58FPQEE244 pKa = 3.71
Molecular weight: 28.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HX16|A0A160HX16_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 1 OX=1843749 PE=4 SV=1
MM1 pKa = 6.86PTVHH5 pKa = 7.6ISEE8 pKa = 4.5TFDD11 pKa = 3.96LSTKK15 pKa = 10.14AGKK18 pKa = 10.16LGLIGIHH25 pKa = 5.47TPPRR29 pKa = 11.84SLVEE33 pKa = 3.74LHH35 pKa = 6.37YY36 pKa = 10.96GPLMSNYY43 pKa = 9.96RR44 pKa = 11.84KK45 pKa = 9.57IRR47 pKa = 11.84FLGADD52 pKa = 3.39VVLACASTLPLDD64 pKa = 3.96PLKK67 pKa = 11.31VGDD70 pKa = 4.21GATLFNPADD79 pKa = 3.53AMNPILYY86 pKa = 9.9KK87 pKa = 10.63AVSNNSFEE95 pKa = 4.27TLVAKK100 pKa = 10.4IKK102 pKa = 10.66GALPVLPSSAGSSIDD117 pKa = 3.12RR118 pKa = 11.84TEE120 pKa = 4.89NIDD123 pKa = 3.72TSTTNYY129 pKa = 10.88DD130 pKa = 3.81DD131 pKa = 4.32FDD133 pKa = 4.09VYY135 pKa = 11.45YY136 pKa = 10.39GILSATFGWKK146 pKa = 10.05KK147 pKa = 11.02SMPQTGLVMRR157 pKa = 11.84NLRR160 pKa = 11.84PLMYY164 pKa = 9.9PLLSDD169 pKa = 3.57IGNDD173 pKa = 2.83IMSASGNIVQDD184 pKa = 3.38STISNNRR191 pKa = 11.84DD192 pKa = 2.97TTSYY196 pKa = 11.03SGIGRR201 pKa = 11.84IARR204 pKa = 11.84GKK206 pKa = 9.28AQRR209 pKa = 11.84APALPLWQGIQSNSGILSPIPSTFPTTYY237 pKa = 8.8VGAVIVPPCHH247 pKa = 6.25TNGSVTYY254 pKa = 9.45YY255 pKa = 10.34RR256 pKa = 11.84LRR258 pKa = 11.84ITWHH262 pKa = 5.47IRR264 pKa = 11.84FEE266 pKa = 4.36GLCPYY271 pKa = 9.57TEE273 pKa = 4.69HH274 pKa = 7.52SPFANFQMAGGATYY288 pKa = 9.09WADD291 pKa = 3.48YY292 pKa = 9.46TGSSKK297 pKa = 10.92DD298 pKa = 3.67FEE300 pKa = 5.58HH301 pKa = 7.89DD302 pKa = 3.35EE303 pKa = 4.59STLSTNGFEE312 pKa = 4.45IEE314 pKa = 4.2KK315 pKa = 10.77VMM317 pKa = 5.07
MM1 pKa = 6.86PTVHH5 pKa = 7.6ISEE8 pKa = 4.5TFDD11 pKa = 3.96LSTKK15 pKa = 10.14AGKK18 pKa = 10.16LGLIGIHH25 pKa = 5.47TPPRR29 pKa = 11.84SLVEE33 pKa = 3.74LHH35 pKa = 6.37YY36 pKa = 10.96GPLMSNYY43 pKa = 9.96RR44 pKa = 11.84KK45 pKa = 9.57IRR47 pKa = 11.84FLGADD52 pKa = 3.39VVLACASTLPLDD64 pKa = 3.96PLKK67 pKa = 11.31VGDD70 pKa = 4.21GATLFNPADD79 pKa = 3.53AMNPILYY86 pKa = 9.9KK87 pKa = 10.63AVSNNSFEE95 pKa = 4.27TLVAKK100 pKa = 10.4IKK102 pKa = 10.66GALPVLPSSAGSSIDD117 pKa = 3.12RR118 pKa = 11.84TEE120 pKa = 4.89NIDD123 pKa = 3.72TSTTNYY129 pKa = 10.88DD130 pKa = 3.81DD131 pKa = 4.32FDD133 pKa = 4.09VYY135 pKa = 11.45YY136 pKa = 10.39GILSATFGWKK146 pKa = 10.05KK147 pKa = 11.02SMPQTGLVMRR157 pKa = 11.84NLRR160 pKa = 11.84PLMYY164 pKa = 9.9PLLSDD169 pKa = 3.57IGNDD173 pKa = 2.83IMSASGNIVQDD184 pKa = 3.38STISNNRR191 pKa = 11.84DD192 pKa = 2.97TTSYY196 pKa = 11.03SGIGRR201 pKa = 11.84IARR204 pKa = 11.84GKK206 pKa = 9.28AQRR209 pKa = 11.84APALPLWQGIQSNSGILSPIPSTFPTTYY237 pKa = 8.8VGAVIVPPCHH247 pKa = 6.25TNGSVTYY254 pKa = 9.45YY255 pKa = 10.34RR256 pKa = 11.84LRR258 pKa = 11.84ITWHH262 pKa = 5.47IRR264 pKa = 11.84FEE266 pKa = 4.36GLCPYY271 pKa = 9.57TEE273 pKa = 4.69HH274 pKa = 7.52SPFANFQMAGGATYY288 pKa = 9.09WADD291 pKa = 3.48YY292 pKa = 9.46TGSSKK297 pKa = 10.92DD298 pKa = 3.67FEE300 pKa = 5.58HH301 pKa = 7.89DD302 pKa = 3.35EE303 pKa = 4.59STLSTNGFEE312 pKa = 4.45IEE314 pKa = 4.2KK315 pKa = 10.77VMM317 pKa = 5.07
Molecular weight: 34.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
561 |
244 |
317 |
280.5 |
31.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.595 ± 0.291 | 1.783 ± 0.707 |
6.061 ± 0.59 | 4.991 ± 1.553 |
4.635 ± 0.718 | 6.952 ± 1.057 |
1.961 ± 0.209 | 6.595 ± 0.291 |
5.704 ± 1.622 | 7.665 ± 0.988 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.743 ± 0.765 | 3.922 ± 0.952 |
6.417 ± 0.442 | 3.03 ± 0.962 |
4.635 ± 0.451 | 7.487 ± 1.939 |
6.595 ± 1.625 | 4.635 ± 0.349 |
2.139 ± 0.742 | 4.456 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
