
Cardiobacteriales bacterium ML27
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cardiobacteriales; Ostreibacteriaceae; Ostreibacterium
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
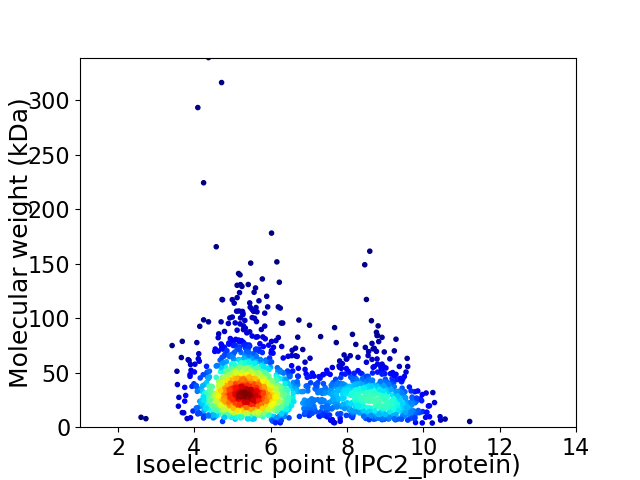
Virtual 2D-PAGE plot for 1905 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
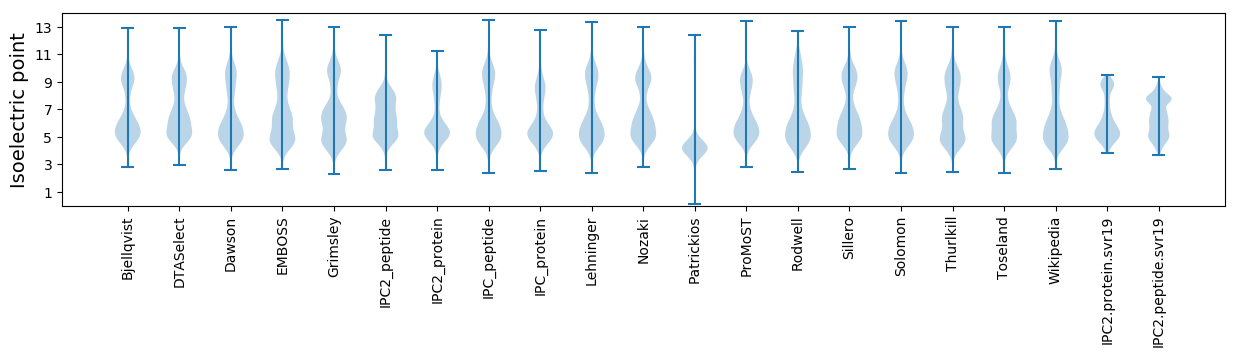
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N7EVF2|A0A6N7EVF2_9GAMM ABC transporter permease subunit OS=Cardiobacteriales bacterium ML27 OX=2654998 GN=GCU85_02345 PE=3 SV=1
MM1 pKa = 7.17YY2 pKa = 10.33RR3 pKa = 11.84SDD5 pKa = 4.2CQNQHH10 pKa = 5.9IMHH13 pKa = 6.39QFAGVFSSRR22 pKa = 11.84AKK24 pKa = 10.22PLVKK28 pKa = 10.26AICLLSGLACTMTIYY43 pKa = 10.5AQEE46 pKa = 4.08VTPVSVNSAGEE57 pKa = 3.85QAASGSYY64 pKa = 8.85WPAISADD71 pKa = 3.09GRR73 pKa = 11.84YY74 pKa = 8.94VAFVSDD80 pKa = 3.76ADD82 pKa = 3.86NLVVGDD88 pKa = 4.27TNGDD92 pKa = 3.42DD93 pKa = 5.5DD94 pKa = 5.15IFVHH98 pKa = 7.08DD99 pKa = 4.84RR100 pKa = 11.84EE101 pKa = 4.56TGEE104 pKa = 4.08TTRR107 pKa = 11.84VSVGSAGEE115 pKa = 3.85QANSQFSAPAISGDD129 pKa = 3.03GRR131 pKa = 11.84YY132 pKa = 8.93VAFISGADD140 pKa = 3.51NLVAGDD146 pKa = 4.12TNNLSDD152 pKa = 3.77IFVHH156 pKa = 7.08DD157 pKa = 4.4RR158 pKa = 11.84EE159 pKa = 4.44TGEE162 pKa = 4.14TNRR165 pKa = 11.84VSVGSAGTQATSGSYY180 pKa = 10.48SPAISADD187 pKa = 2.84GRR189 pKa = 11.84YY190 pKa = 8.94VAFEE194 pKa = 3.88SWADD198 pKa = 3.52NLVAGDD204 pKa = 4.14TNNEE208 pKa = 3.51SDD210 pKa = 3.58IFVHH214 pKa = 7.27DD215 pKa = 4.4RR216 pKa = 11.84EE217 pKa = 4.4TDD219 pKa = 2.91EE220 pKa = 4.2TTRR223 pKa = 11.84VSVSSAGTQANGGSEE238 pKa = 4.26SPAISADD245 pKa = 2.89GRR247 pKa = 11.84YY248 pKa = 8.88VAFEE252 pKa = 4.03SWVDD256 pKa = 3.4NLVAGDD262 pKa = 3.8TNNRR266 pKa = 11.84TDD268 pKa = 3.1IFVHH272 pKa = 6.4DD273 pKa = 4.57RR274 pKa = 11.84EE275 pKa = 4.56TGEE278 pKa = 4.08TTRR281 pKa = 11.84VSVGSAGEE289 pKa = 3.85QANSQFSAPAISGDD303 pKa = 3.03GRR305 pKa = 11.84YY306 pKa = 8.95VAFVSSADD314 pKa = 3.58NLVAGDD320 pKa = 3.78TTNNNWDD327 pKa = 3.45VFVRR331 pKa = 11.84NLEE334 pKa = 3.87TGEE337 pKa = 4.16IKK339 pKa = 10.65RR340 pKa = 11.84ITADD344 pKa = 2.84IPYY347 pKa = 9.81EE348 pKa = 4.06EE349 pKa = 5.25EE350 pKa = 3.65ISSAVYY356 pKa = 10.47YY357 pKa = 10.18RR358 pKa = 11.84VSLSADD364 pKa = 2.87GRR366 pKa = 11.84YY367 pKa = 8.96IAYY370 pKa = 10.04SPEE373 pKa = 3.73YY374 pKa = 9.27QACDD378 pKa = 3.61FLGCGPSFYY387 pKa = 10.7GIYY390 pKa = 9.92IYY392 pKa = 10.6DD393 pKa = 3.63QEE395 pKa = 4.49TGQTEE400 pKa = 4.52VIAALLDD407 pKa = 3.79EE408 pKa = 4.86NNEE411 pKa = 4.52FGNSWPYY418 pKa = 10.5NGSGFPSLSADD429 pKa = 3.39GQTVAFGSSATNLVPNDD446 pKa = 3.48TNEE449 pKa = 3.84AFDD452 pKa = 3.74IFVAEE457 pKa = 4.23LGEE460 pKa = 4.42TISNISGTWYY470 pKa = 10.3DD471 pKa = 3.88RR472 pKa = 11.84NQSGG476 pKa = 3.81
MM1 pKa = 7.17YY2 pKa = 10.33RR3 pKa = 11.84SDD5 pKa = 4.2CQNQHH10 pKa = 5.9IMHH13 pKa = 6.39QFAGVFSSRR22 pKa = 11.84AKK24 pKa = 10.22PLVKK28 pKa = 10.26AICLLSGLACTMTIYY43 pKa = 10.5AQEE46 pKa = 4.08VTPVSVNSAGEE57 pKa = 3.85QAASGSYY64 pKa = 8.85WPAISADD71 pKa = 3.09GRR73 pKa = 11.84YY74 pKa = 8.94VAFVSDD80 pKa = 3.76ADD82 pKa = 3.86NLVVGDD88 pKa = 4.27TNGDD92 pKa = 3.42DD93 pKa = 5.5DD94 pKa = 5.15IFVHH98 pKa = 7.08DD99 pKa = 4.84RR100 pKa = 11.84EE101 pKa = 4.56TGEE104 pKa = 4.08TTRR107 pKa = 11.84VSVGSAGEE115 pKa = 3.85QANSQFSAPAISGDD129 pKa = 3.03GRR131 pKa = 11.84YY132 pKa = 8.93VAFISGADD140 pKa = 3.51NLVAGDD146 pKa = 4.12TNNLSDD152 pKa = 3.77IFVHH156 pKa = 7.08DD157 pKa = 4.4RR158 pKa = 11.84EE159 pKa = 4.44TGEE162 pKa = 4.14TNRR165 pKa = 11.84VSVGSAGTQATSGSYY180 pKa = 10.48SPAISADD187 pKa = 2.84GRR189 pKa = 11.84YY190 pKa = 8.94VAFEE194 pKa = 3.88SWADD198 pKa = 3.52NLVAGDD204 pKa = 4.14TNNEE208 pKa = 3.51SDD210 pKa = 3.58IFVHH214 pKa = 7.27DD215 pKa = 4.4RR216 pKa = 11.84EE217 pKa = 4.4TDD219 pKa = 2.91EE220 pKa = 4.2TTRR223 pKa = 11.84VSVSSAGTQANGGSEE238 pKa = 4.26SPAISADD245 pKa = 2.89GRR247 pKa = 11.84YY248 pKa = 8.88VAFEE252 pKa = 4.03SWVDD256 pKa = 3.4NLVAGDD262 pKa = 3.8TNNRR266 pKa = 11.84TDD268 pKa = 3.1IFVHH272 pKa = 6.4DD273 pKa = 4.57RR274 pKa = 11.84EE275 pKa = 4.56TGEE278 pKa = 4.08TTRR281 pKa = 11.84VSVGSAGEE289 pKa = 3.85QANSQFSAPAISGDD303 pKa = 3.03GRR305 pKa = 11.84YY306 pKa = 8.95VAFVSSADD314 pKa = 3.58NLVAGDD320 pKa = 3.78TTNNNWDD327 pKa = 3.45VFVRR331 pKa = 11.84NLEE334 pKa = 3.87TGEE337 pKa = 4.16IKK339 pKa = 10.65RR340 pKa = 11.84ITADD344 pKa = 2.84IPYY347 pKa = 9.81EE348 pKa = 4.06EE349 pKa = 5.25EE350 pKa = 3.65ISSAVYY356 pKa = 10.47YY357 pKa = 10.18RR358 pKa = 11.84VSLSADD364 pKa = 2.87GRR366 pKa = 11.84YY367 pKa = 8.96IAYY370 pKa = 10.04SPEE373 pKa = 3.73YY374 pKa = 9.27QACDD378 pKa = 3.61FLGCGPSFYY387 pKa = 10.7GIYY390 pKa = 9.92IYY392 pKa = 10.6DD393 pKa = 3.63QEE395 pKa = 4.49TGQTEE400 pKa = 4.52VIAALLDD407 pKa = 3.79EE408 pKa = 4.86NNEE411 pKa = 4.52FGNSWPYY418 pKa = 10.5NGSGFPSLSADD429 pKa = 3.39GQTVAFGSSATNLVPNDD446 pKa = 3.48TNEE449 pKa = 3.84AFDD452 pKa = 3.74IFVAEE457 pKa = 4.23LGEE460 pKa = 4.42TISNISGTWYY470 pKa = 10.3DD471 pKa = 3.88RR472 pKa = 11.84NQSGG476 pKa = 3.81
Molecular weight: 50.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N7EVQ6|A0A6N7EVQ6_9GAMM Uncharacterized protein OS=Cardiobacteriales bacterium ML27 OX=2654998 GN=GCU85_00220 PE=4 SV=1
MM1 pKa = 6.65KK2 pKa = 10.14QKK4 pKa = 9.7RR5 pKa = 11.84TFQPSNIKK13 pKa = 10.11RR14 pKa = 11.84KK15 pKa = 9.04RR16 pKa = 11.84SHH18 pKa = 6.18GFRR21 pKa = 11.84ARR23 pKa = 11.84MATAGGRR30 pKa = 11.84AVINARR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.67GRR41 pKa = 11.84KK42 pKa = 8.91RR43 pKa = 11.84LSAA46 pKa = 3.96
MM1 pKa = 6.65KK2 pKa = 10.14QKK4 pKa = 9.7RR5 pKa = 11.84TFQPSNIKK13 pKa = 10.11RR14 pKa = 11.84KK15 pKa = 9.04RR16 pKa = 11.84SHH18 pKa = 6.18GFRR21 pKa = 11.84ARR23 pKa = 11.84MATAGGRR30 pKa = 11.84AVINARR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.67GRR41 pKa = 11.84KK42 pKa = 8.91RR43 pKa = 11.84LSAA46 pKa = 3.96
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
628582 |
29 |
3124 |
330.0 |
36.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.821 ± 0.071 | 1.005 ± 0.02 |
5.669 ± 0.049 | 5.17 ± 0.056 |
4.007 ± 0.04 | 6.677 ± 0.055 |
2.315 ± 0.031 | 6.979 ± 0.047 |
4.852 ± 0.057 | 10.018 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.026 | 4.378 ± 0.044 |
4.146 ± 0.036 | 4.882 ± 0.052 |
4.58 ± 0.034 | 6.133 ± 0.045 |
6.165 ± 0.047 | 6.578 ± 0.047 |
1.107 ± 0.021 | 3.115 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
