
Butyrivibrio proteoclasticus (strain ATCC 51982 / DSM 14932 / B316) (Clostridium proteoclasticum)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio; Butyrivibrio proteoclasticus
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
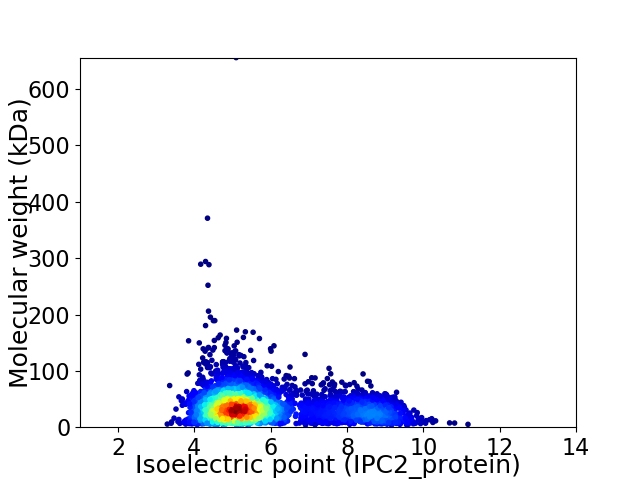
Virtual 2D-PAGE plot for 3799 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0RZY4|E0RZY4_BUTPB Acyltransferase OS=Butyrivibrio proteoclasticus (strain ATCC 51982 / DSM 14932 / B316) OX=515622 GN=bpr_I0437 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 10.42KK3 pKa = 10.14KK4 pKa = 10.78LSVLLAGVMVCTMFTACGKK23 pKa = 10.25EE24 pKa = 4.05NASEE28 pKa = 4.08NADD31 pKa = 3.31NAASTEE37 pKa = 3.87NSADD41 pKa = 3.33AAATEE46 pKa = 4.27TSVAEE51 pKa = 4.59GMALTIDD58 pKa = 4.19FDD60 pKa = 4.1NLEE63 pKa = 4.43TEE65 pKa = 4.34TLNDD69 pKa = 3.24VTASDD74 pKa = 3.86IVTLGQYY81 pKa = 10.14KK82 pKa = 9.97EE83 pKa = 4.35LSFEE87 pKa = 4.14VAKK90 pKa = 11.18DD91 pKa = 3.57EE92 pKa = 4.36VTDD95 pKa = 4.67AEE97 pKa = 4.45VEE99 pKa = 4.35DD100 pKa = 4.44YY101 pKa = 10.91VANLWSTKK109 pKa = 10.35PLMLDD114 pKa = 3.2VTDD117 pKa = 5.0RR118 pKa = 11.84AVQDD122 pKa = 3.49GDD124 pKa = 4.11TVNIDD129 pKa = 3.63YY130 pKa = 10.54VGKK133 pKa = 10.53YY134 pKa = 10.47ADD136 pKa = 3.68TKK138 pKa = 10.45EE139 pKa = 4.28AFDD142 pKa = 3.99GGTAEE147 pKa = 4.6GASLTIGSNSYY158 pKa = 10.55IEE160 pKa = 4.6GFEE163 pKa = 4.17SGLVGVNIGDD173 pKa = 3.92TVDD176 pKa = 4.04LNLTFPEE183 pKa = 4.25NYY185 pKa = 9.72GAEE188 pKa = 4.06NLAGKK193 pKa = 10.07DD194 pKa = 3.3VVFTVTVNSIQAPDD208 pKa = 3.6TEE210 pKa = 5.27MSDD213 pKa = 2.92EE214 pKa = 3.93WAAGLGYY221 pKa = 10.82DD222 pKa = 4.84GVTDD226 pKa = 4.6LATLRR231 pKa = 11.84DD232 pKa = 4.0YY233 pKa = 11.38ARR235 pKa = 11.84KK236 pKa = 7.73TLEE239 pKa = 4.24DD240 pKa = 3.27QAQNEE245 pKa = 4.24FDD247 pKa = 3.7STVEE251 pKa = 3.8NTAVQTVYY259 pKa = 11.22DD260 pKa = 3.85STAFGDD266 pKa = 3.83IPQALINRR274 pKa = 11.84YY275 pKa = 7.78MKK277 pKa = 9.8QQKK280 pKa = 9.89QMLDD284 pKa = 3.66YY285 pKa = 10.4QATMYY290 pKa = 10.38SYY292 pKa = 11.61YY293 pKa = 10.63YY294 pKa = 9.95GQQLTADD301 pKa = 4.0SLLANYY307 pKa = 8.23MNAEE311 pKa = 4.15GFVGTVDD318 pKa = 5.59DD319 pKa = 4.35YY320 pKa = 12.15LNDD323 pKa = 3.31ISKK326 pKa = 11.13DD327 pKa = 3.29MAQQYY332 pKa = 11.55LMFQAIADD340 pKa = 4.06EE341 pKa = 4.39EE342 pKa = 4.67NITVSDD348 pKa = 4.12EE349 pKa = 4.51EE350 pKa = 3.75IDD352 pKa = 4.53AYY354 pKa = 11.15LKK356 pKa = 10.55NAYY359 pKa = 8.9EE360 pKa = 4.23NASTTSFSSFEE371 pKa = 4.14EE372 pKa = 4.31YY373 pKa = 10.41KK374 pKa = 10.82EE375 pKa = 4.16SLDD378 pKa = 3.92LEE380 pKa = 4.66IYY382 pKa = 10.59RR383 pKa = 11.84EE384 pKa = 3.83GLMADD389 pKa = 3.38KK390 pKa = 10.85VVAFIVEE397 pKa = 4.17NANVVATAATEE408 pKa = 4.09DD409 pKa = 3.75AASEE413 pKa = 4.28SSSAEE418 pKa = 3.63ASADD422 pKa = 3.44AASEE426 pKa = 4.24SSSTEE431 pKa = 3.88TVAEE435 pKa = 4.14
MM1 pKa = 7.4KK2 pKa = 10.42KK3 pKa = 10.14KK4 pKa = 10.78LSVLLAGVMVCTMFTACGKK23 pKa = 10.25EE24 pKa = 4.05NASEE28 pKa = 4.08NADD31 pKa = 3.31NAASTEE37 pKa = 3.87NSADD41 pKa = 3.33AAATEE46 pKa = 4.27TSVAEE51 pKa = 4.59GMALTIDD58 pKa = 4.19FDD60 pKa = 4.1NLEE63 pKa = 4.43TEE65 pKa = 4.34TLNDD69 pKa = 3.24VTASDD74 pKa = 3.86IVTLGQYY81 pKa = 10.14KK82 pKa = 9.97EE83 pKa = 4.35LSFEE87 pKa = 4.14VAKK90 pKa = 11.18DD91 pKa = 3.57EE92 pKa = 4.36VTDD95 pKa = 4.67AEE97 pKa = 4.45VEE99 pKa = 4.35DD100 pKa = 4.44YY101 pKa = 10.91VANLWSTKK109 pKa = 10.35PLMLDD114 pKa = 3.2VTDD117 pKa = 5.0RR118 pKa = 11.84AVQDD122 pKa = 3.49GDD124 pKa = 4.11TVNIDD129 pKa = 3.63YY130 pKa = 10.54VGKK133 pKa = 10.53YY134 pKa = 10.47ADD136 pKa = 3.68TKK138 pKa = 10.45EE139 pKa = 4.28AFDD142 pKa = 3.99GGTAEE147 pKa = 4.6GASLTIGSNSYY158 pKa = 10.55IEE160 pKa = 4.6GFEE163 pKa = 4.17SGLVGVNIGDD173 pKa = 3.92TVDD176 pKa = 4.04LNLTFPEE183 pKa = 4.25NYY185 pKa = 9.72GAEE188 pKa = 4.06NLAGKK193 pKa = 10.07DD194 pKa = 3.3VVFTVTVNSIQAPDD208 pKa = 3.6TEE210 pKa = 5.27MSDD213 pKa = 2.92EE214 pKa = 3.93WAAGLGYY221 pKa = 10.82DD222 pKa = 4.84GVTDD226 pKa = 4.6LATLRR231 pKa = 11.84DD232 pKa = 4.0YY233 pKa = 11.38ARR235 pKa = 11.84KK236 pKa = 7.73TLEE239 pKa = 4.24DD240 pKa = 3.27QAQNEE245 pKa = 4.24FDD247 pKa = 3.7STVEE251 pKa = 3.8NTAVQTVYY259 pKa = 11.22DD260 pKa = 3.85STAFGDD266 pKa = 3.83IPQALINRR274 pKa = 11.84YY275 pKa = 7.78MKK277 pKa = 9.8QQKK280 pKa = 9.89QMLDD284 pKa = 3.66YY285 pKa = 10.4QATMYY290 pKa = 10.38SYY292 pKa = 11.61YY293 pKa = 10.63YY294 pKa = 9.95GQQLTADD301 pKa = 4.0SLLANYY307 pKa = 8.23MNAEE311 pKa = 4.15GFVGTVDD318 pKa = 5.59DD319 pKa = 4.35YY320 pKa = 12.15LNDD323 pKa = 3.31ISKK326 pKa = 11.13DD327 pKa = 3.29MAQQYY332 pKa = 11.55LMFQAIADD340 pKa = 4.06EE341 pKa = 4.39EE342 pKa = 4.67NITVSDD348 pKa = 4.12EE349 pKa = 4.51EE350 pKa = 3.75IDD352 pKa = 4.53AYY354 pKa = 11.15LKK356 pKa = 10.55NAYY359 pKa = 8.9EE360 pKa = 4.23NASTTSFSSFEE371 pKa = 4.14EE372 pKa = 4.31YY373 pKa = 10.41KK374 pKa = 10.82EE375 pKa = 4.16SLDD378 pKa = 3.92LEE380 pKa = 4.66IYY382 pKa = 10.59RR383 pKa = 11.84EE384 pKa = 3.83GLMADD389 pKa = 3.38KK390 pKa = 10.85VVAFIVEE397 pKa = 4.17NANVVATAATEE408 pKa = 4.09DD409 pKa = 3.75AASEE413 pKa = 4.28SSSAEE418 pKa = 3.63ASADD422 pKa = 3.44AASEE426 pKa = 4.24SSSTEE431 pKa = 3.88TVAEE435 pKa = 4.14
Molecular weight: 47.05 kDa
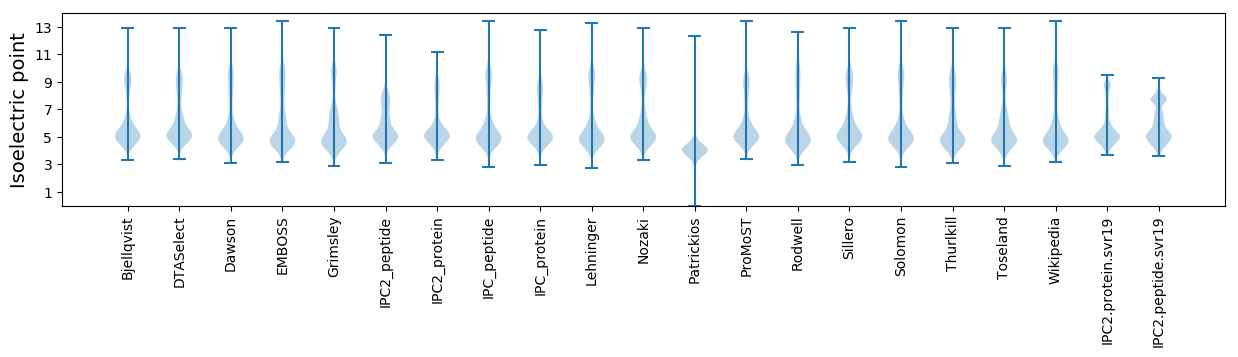
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0RW10|E0RW10_BUTPB AAA-ATPase_like domain-containing protein OS=Butyrivibrio proteoclasticus (strain ATCC 51982 / DSM 14932 / B316) OX=515622 GN=bpr_I0125 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1301855 |
30 |
5853 |
342.7 |
38.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.2 ± 0.046 | 1.373 ± 0.017 |
6.63 ± 0.033 | 7.243 ± 0.042 |
4.317 ± 0.03 | 6.819 ± 0.039 |
1.635 ± 0.015 | 7.696 ± 0.041 |
7.02 ± 0.036 | 8.329 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.993 ± 0.02 | 4.905 ± 0.03 |
3.079 ± 0.022 | 2.76 ± 0.019 |
4.064 ± 0.029 | 6.279 ± 0.032 |
5.345 ± 0.039 | 6.777 ± 0.028 |
0.938 ± 0.016 | 4.597 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
