
Cyclovirus ZM32
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Human associated cyclovirus 8; Cyclovirus VN
Average proteome isoelectric point is 8.45
Get precalculated fractions of proteins
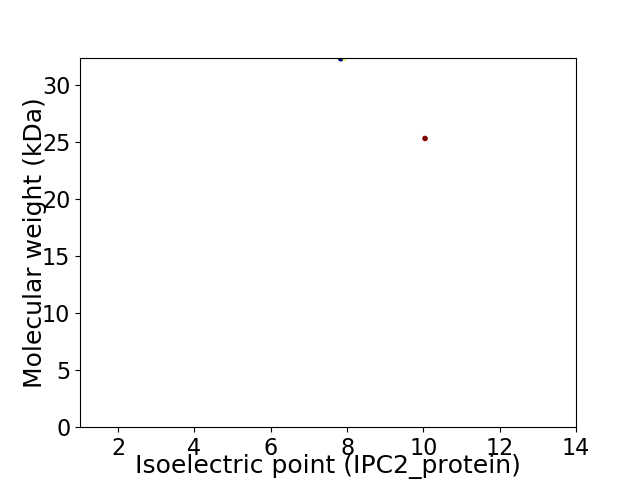
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097ZPG6|A0A097ZPG6_9CIRC ATP-dependent helicase Rep OS=Cyclovirus ZM32 OX=1506566 GN=Rep PE=3 SV=1
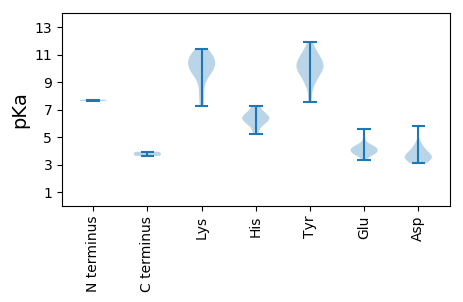
MM1 pKa = 7.72ANNSTVRR8 pKa = 11.84RR9 pKa = 11.84FCWTLNNYY17 pKa = 8.05TDD19 pKa = 3.69EE20 pKa = 5.01DD21 pKa = 4.2VDD23 pKa = 4.45VLQKK27 pKa = 11.03DD28 pKa = 3.5LAEE31 pKa = 4.3LCKK34 pKa = 10.04FAIFGRR40 pKa = 11.84EE41 pKa = 3.98TCPSTGTKK49 pKa = 9.84HH50 pKa = 6.5LQGFCNLQRR59 pKa = 11.84PKK61 pKa = 10.35RR62 pKa = 11.84FNSIRR67 pKa = 11.84QIFGGRR73 pKa = 11.84AHH75 pKa = 7.27IEE77 pKa = 3.81RR78 pKa = 11.84AKK80 pKa = 11.13GSDD83 pKa = 3.64LEE85 pKa = 4.17NQEE88 pKa = 4.26YY89 pKa = 10.11CSKK92 pKa = 11.24GGDD95 pKa = 3.09VWQCGSPSTQGTRR108 pKa = 11.84TDD110 pKa = 3.47LQKK113 pKa = 10.72VVSVIEE119 pKa = 4.21GGEE122 pKa = 3.88RR123 pKa = 11.84NIKK126 pKa = 10.07NLALQFPTAYY136 pKa = 9.14IKK138 pKa = 10.39YY139 pKa = 9.66FKK141 pKa = 10.74GIEE144 pKa = 3.78QYY146 pKa = 10.83IRR148 pKa = 11.84IAHH151 pKa = 6.42GNPEE155 pKa = 4.25RR156 pKa = 11.84DD157 pKa = 3.42FKK159 pKa = 11.37TEE161 pKa = 3.7VFYY164 pKa = 10.55FWGATGTGKK173 pKa = 9.99SRR175 pKa = 11.84KK176 pKa = 9.34ARR178 pKa = 11.84EE179 pKa = 3.79EE180 pKa = 3.77SRR182 pKa = 11.84ALGDD186 pKa = 3.09TYY188 pKa = 11.22YY189 pKa = 10.96KK190 pKa = 10.5PRR192 pKa = 11.84GEE194 pKa = 3.75WWDD197 pKa = 3.93GYY199 pKa = 9.14TGQPCVVIDD208 pKa = 5.82DD209 pKa = 4.49FYY211 pKa = 11.88GWIKK215 pKa = 10.75YY216 pKa = 10.28DD217 pKa = 4.1EE218 pKa = 4.44LLKK221 pKa = 10.71ICDD224 pKa = 3.7RR225 pKa = 11.84YY226 pKa = 10.06PYY228 pKa = 9.86RR229 pKa = 11.84VPVKK233 pKa = 10.11GGYY236 pKa = 9.35EE237 pKa = 4.03NFVSKK242 pKa = 9.88YY243 pKa = 8.77IWITSEE249 pKa = 3.94RR250 pKa = 11.84PLEE253 pKa = 3.78EE254 pKa = 3.95VYY256 pKa = 10.77RR257 pKa = 11.84FLAYY261 pKa = 10.18DD262 pKa = 3.7CSSLRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84LNKK272 pKa = 9.94EE273 pKa = 3.29IHH275 pKa = 6.23FEE277 pKa = 3.93
MM1 pKa = 7.72ANNSTVRR8 pKa = 11.84RR9 pKa = 11.84FCWTLNNYY17 pKa = 8.05TDD19 pKa = 3.69EE20 pKa = 5.01DD21 pKa = 4.2VDD23 pKa = 4.45VLQKK27 pKa = 11.03DD28 pKa = 3.5LAEE31 pKa = 4.3LCKK34 pKa = 10.04FAIFGRR40 pKa = 11.84EE41 pKa = 3.98TCPSTGTKK49 pKa = 9.84HH50 pKa = 6.5LQGFCNLQRR59 pKa = 11.84PKK61 pKa = 10.35RR62 pKa = 11.84FNSIRR67 pKa = 11.84QIFGGRR73 pKa = 11.84AHH75 pKa = 7.27IEE77 pKa = 3.81RR78 pKa = 11.84AKK80 pKa = 11.13GSDD83 pKa = 3.64LEE85 pKa = 4.17NQEE88 pKa = 4.26YY89 pKa = 10.11CSKK92 pKa = 11.24GGDD95 pKa = 3.09VWQCGSPSTQGTRR108 pKa = 11.84TDD110 pKa = 3.47LQKK113 pKa = 10.72VVSVIEE119 pKa = 4.21GGEE122 pKa = 3.88RR123 pKa = 11.84NIKK126 pKa = 10.07NLALQFPTAYY136 pKa = 9.14IKK138 pKa = 10.39YY139 pKa = 9.66FKK141 pKa = 10.74GIEE144 pKa = 3.78QYY146 pKa = 10.83IRR148 pKa = 11.84IAHH151 pKa = 6.42GNPEE155 pKa = 4.25RR156 pKa = 11.84DD157 pKa = 3.42FKK159 pKa = 11.37TEE161 pKa = 3.7VFYY164 pKa = 10.55FWGATGTGKK173 pKa = 9.99SRR175 pKa = 11.84KK176 pKa = 9.34ARR178 pKa = 11.84EE179 pKa = 3.79EE180 pKa = 3.77SRR182 pKa = 11.84ALGDD186 pKa = 3.09TYY188 pKa = 11.22YY189 pKa = 10.96KK190 pKa = 10.5PRR192 pKa = 11.84GEE194 pKa = 3.75WWDD197 pKa = 3.93GYY199 pKa = 9.14TGQPCVVIDD208 pKa = 5.82DD209 pKa = 4.49FYY211 pKa = 11.88GWIKK215 pKa = 10.75YY216 pKa = 10.28DD217 pKa = 4.1EE218 pKa = 4.44LLKK221 pKa = 10.71ICDD224 pKa = 3.7RR225 pKa = 11.84YY226 pKa = 10.06PYY228 pKa = 9.86RR229 pKa = 11.84VPVKK233 pKa = 10.11GGYY236 pKa = 9.35EE237 pKa = 4.03NFVSKK242 pKa = 9.88YY243 pKa = 8.77IWITSEE249 pKa = 3.94RR250 pKa = 11.84PLEE253 pKa = 3.78EE254 pKa = 3.95VYY256 pKa = 10.77RR257 pKa = 11.84FLAYY261 pKa = 10.18DD262 pKa = 3.7CSSLRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84LNKK272 pKa = 9.94EE273 pKa = 3.29IHH275 pKa = 6.23FEE277 pKa = 3.93
Molecular weight: 32.31 kDa
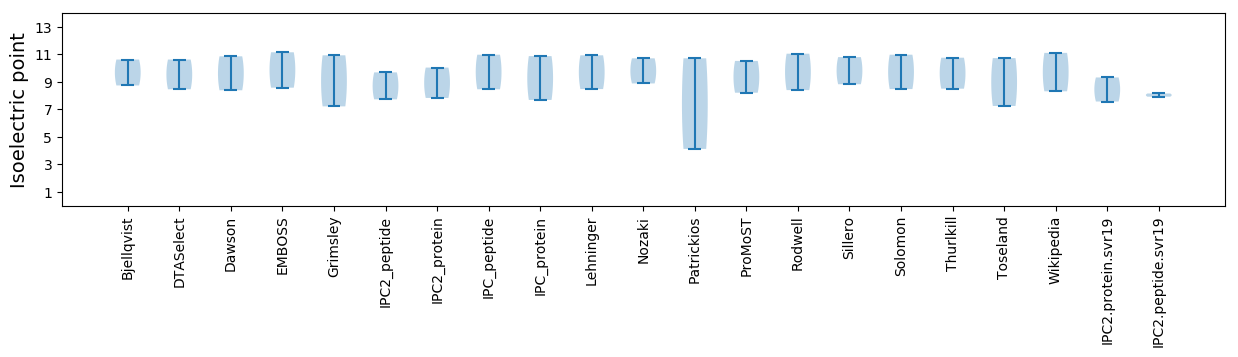
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097ZPG6|A0A097ZPG6_9CIRC ATP-dependent helicase Rep OS=Cyclovirus ZM32 OX=1506566 GN=Rep PE=3 SV=1
MM1 pKa = 7.62AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84VIRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84APPRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ATIRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84KK28 pKa = 8.17KK29 pKa = 7.23TFRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.26TGNFTLKK41 pKa = 9.94VRR43 pKa = 11.84KK44 pKa = 7.58TAVLAIDD51 pKa = 3.4SDD53 pKa = 3.25KK54 pKa = 11.3GYY56 pKa = 10.16SYY58 pKa = 11.61NVAPTLNDD66 pKa = 3.25YY67 pKa = 11.21EE68 pKa = 4.26EE69 pKa = 4.46AKK71 pKa = 10.53PYY73 pKa = 10.17IPYY76 pKa = 10.15FEE78 pKa = 5.6SYY80 pKa = 10.47RR81 pKa = 11.84IWNCSVKK88 pKa = 9.12ITPLFNVASPGEE100 pKa = 4.16AVPRR104 pKa = 11.84YY105 pKa = 9.83YY106 pKa = 10.63SAPWHH111 pKa = 6.06RR112 pKa = 11.84AGPAALSTNSILSLDD127 pKa = 3.56RR128 pKa = 11.84AKK130 pKa = 10.81SHH132 pKa = 6.64NGTSGSFRR140 pKa = 11.84RR141 pKa = 11.84YY142 pKa = 7.53TPCILTDD149 pKa = 3.39TSVAGTPGQKK159 pKa = 9.62LGKK162 pKa = 9.05MDD164 pKa = 3.94WRR166 pKa = 11.84PKK168 pKa = 9.9IALQVSSTAIPHH180 pKa = 5.19YY181 pKa = 9.82CALYY185 pKa = 9.66HH186 pKa = 6.29WSIDD190 pKa = 3.63QLQGSQITQRR200 pKa = 11.84QYY202 pKa = 10.86EE203 pKa = 4.21IEE205 pKa = 4.13IMSTITFYY213 pKa = 9.05TQKK216 pKa = 10.79SFAGG220 pKa = 3.62
MM1 pKa = 7.62AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84VIRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84APPRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ATIRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84LRR27 pKa = 11.84KK28 pKa = 8.17KK29 pKa = 7.23TFRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.26TGNFTLKK41 pKa = 9.94VRR43 pKa = 11.84KK44 pKa = 7.58TAVLAIDD51 pKa = 3.4SDD53 pKa = 3.25KK54 pKa = 11.3GYY56 pKa = 10.16SYY58 pKa = 11.61NVAPTLNDD66 pKa = 3.25YY67 pKa = 11.21EE68 pKa = 4.26EE69 pKa = 4.46AKK71 pKa = 10.53PYY73 pKa = 10.17IPYY76 pKa = 10.15FEE78 pKa = 5.6SYY80 pKa = 10.47RR81 pKa = 11.84IWNCSVKK88 pKa = 9.12ITPLFNVASPGEE100 pKa = 4.16AVPRR104 pKa = 11.84YY105 pKa = 9.83YY106 pKa = 10.63SAPWHH111 pKa = 6.06RR112 pKa = 11.84AGPAALSTNSILSLDD127 pKa = 3.56RR128 pKa = 11.84AKK130 pKa = 10.81SHH132 pKa = 6.64NGTSGSFRR140 pKa = 11.84RR141 pKa = 11.84YY142 pKa = 7.53TPCILTDD149 pKa = 3.39TSVAGTPGQKK159 pKa = 9.62LGKK162 pKa = 9.05MDD164 pKa = 3.94WRR166 pKa = 11.84PKK168 pKa = 9.9IALQVSSTAIPHH180 pKa = 5.19YY181 pKa = 9.82CALYY185 pKa = 9.66HH186 pKa = 6.29WSIDD190 pKa = 3.63QLQGSQITQRR200 pKa = 11.84QYY202 pKa = 10.86EE203 pKa = 4.21IEE205 pKa = 4.13IMSTITFYY213 pKa = 9.05TQKK216 pKa = 10.79SFAGG220 pKa = 3.62
Molecular weight: 25.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
497 |
220 |
277 |
248.5 |
28.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.237 ± 1.578 | 2.414 ± 0.691 |
4.427 ± 0.819 | 5.433 ± 1.779 |
4.628 ± 0.652 | 7.042 ± 1.343 |
1.61 ± 0.137 | 6.237 ± 0.382 |
6.439 ± 0.348 | 6.036 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.805 ± 0.368 | 3.823 ± 0.422 |
4.829 ± 1.009 | 3.823 ± 0.123 |
9.859 ± 1.288 | 6.64 ± 1.313 |
6.841 ± 0.882 | 4.628 ± 0.353 |
2.213 ± 0.26 | 6.036 ± 0.084 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
