
Torque teno midi virus 8
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
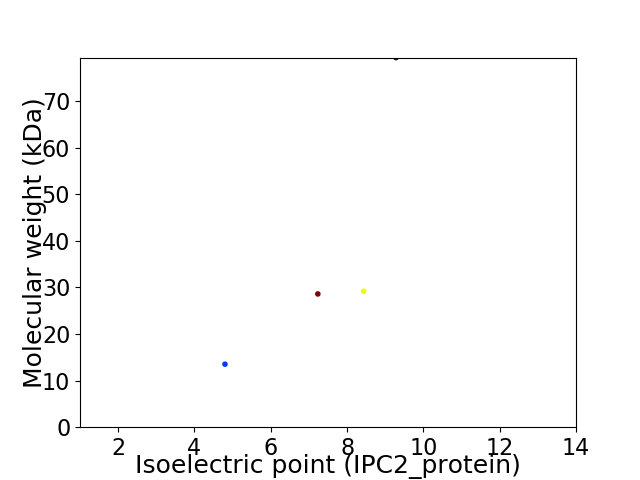
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A7VLY0|A7VLY0_9VIRU Capsid protein OS=Torque teno midi virus 8 OX=2065049 PE=3 SV=1
MM1 pKa = 7.16QHH3 pKa = 6.75LCSKK7 pKa = 10.96DD8 pKa = 3.54FIKK11 pKa = 10.37PCKK14 pKa = 9.98YY15 pKa = 10.54NEE17 pKa = 3.98EE18 pKa = 4.39TKK20 pKa = 10.6NQLWMSEE27 pKa = 4.22LADD30 pKa = 4.65SHH32 pKa = 7.26DD33 pKa = 3.69IWCSCDD39 pKa = 3.08TPFAHH44 pKa = 7.44LLASIFPPGHH54 pKa = 5.56QDD56 pKa = 3.05RR57 pKa = 11.84GKK59 pKa = 8.84TIQDD63 pKa = 3.04ILEE66 pKa = 4.11RR67 pKa = 11.84DD68 pKa = 4.05YY69 pKa = 11.58KK70 pKa = 9.77QLCHH74 pKa = 6.65SGGPAEE80 pKa = 4.34RR81 pKa = 11.84SHH83 pKa = 7.38GILTGTATGADD94 pKa = 3.25IKK96 pKa = 11.27EE97 pKa = 4.02EE98 pKa = 4.58DD99 pKa = 4.14GLEE102 pKa = 4.15GDD104 pKa = 4.02AEE106 pKa = 4.39EE107 pKa = 4.93FLMAAAAAAAEE118 pKa = 4.39EE119 pKa = 4.57KK120 pKa = 10.61EE121 pKa = 4.23RR122 pKa = 4.7
MM1 pKa = 7.16QHH3 pKa = 6.75LCSKK7 pKa = 10.96DD8 pKa = 3.54FIKK11 pKa = 10.37PCKK14 pKa = 9.98YY15 pKa = 10.54NEE17 pKa = 3.98EE18 pKa = 4.39TKK20 pKa = 10.6NQLWMSEE27 pKa = 4.22LADD30 pKa = 4.65SHH32 pKa = 7.26DD33 pKa = 3.69IWCSCDD39 pKa = 3.08TPFAHH44 pKa = 7.44LLASIFPPGHH54 pKa = 5.56QDD56 pKa = 3.05RR57 pKa = 11.84GKK59 pKa = 8.84TIQDD63 pKa = 3.04ILEE66 pKa = 4.11RR67 pKa = 11.84DD68 pKa = 4.05YY69 pKa = 11.58KK70 pKa = 9.77QLCHH74 pKa = 6.65SGGPAEE80 pKa = 4.34RR81 pKa = 11.84SHH83 pKa = 7.38GILTGTATGADD94 pKa = 3.25IKK96 pKa = 11.27EE97 pKa = 4.02EE98 pKa = 4.58DD99 pKa = 4.14GLEE102 pKa = 4.15GDD104 pKa = 4.02AEE106 pKa = 4.39EE107 pKa = 4.93FLMAAAAAAAEE118 pKa = 4.39EE119 pKa = 4.57KK120 pKa = 10.61EE121 pKa = 4.23RR122 pKa = 4.7
Molecular weight: 13.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLY0|A7VLY0_9VIRU Capsid protein OS=Torque teno midi virus 8 OX=2065049 PE=3 SV=1
MM1 pKa = 7.31PFWWARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.25KK10 pKa = 9.8PWYY13 pKa = 9.88SNWNRR18 pKa = 11.84YY19 pKa = 6.51RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 8.8KK24 pKa = 9.61RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84WPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84IPYY38 pKa = 7.85GRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 8.72RR49 pKa = 11.84KK50 pKa = 8.81VRR52 pKa = 11.84RR53 pKa = 11.84KK54 pKa = 7.41RR55 pKa = 11.84QRR57 pKa = 11.84IIVKK61 pKa = 8.67QWQPEE66 pKa = 4.43SIVKK70 pKa = 10.5CEE72 pKa = 3.87IKK74 pKa = 10.97GLGCLVAGAQGRR86 pKa = 11.84QPFCYY91 pKa = 9.55TNSKK95 pKa = 10.36YY96 pKa = 10.54EE97 pKa = 3.96YY98 pKa = 8.79PQPKK102 pKa = 10.0APGGGGFGAEE112 pKa = 4.52LFTLQYY118 pKa = 11.06LYY120 pKa = 10.94DD121 pKa = 3.44EE122 pKa = 4.42WVARR126 pKa = 11.84RR127 pKa = 11.84NIWTKK132 pKa = 9.79TNDD135 pKa = 3.36YY136 pKa = 10.38KK137 pKa = 11.52DD138 pKa = 3.57LVRR141 pKa = 11.84FTGATFYY148 pKa = 10.72LYY150 pKa = 10.58KK151 pKa = 10.68HH152 pKa = 6.27PTTDD156 pKa = 5.35FIFNYY161 pKa = 10.01NNQPPFLLEE170 pKa = 3.59KK171 pKa = 10.73DD172 pKa = 4.62FYY174 pKa = 11.67NDD176 pKa = 3.27LHH178 pKa = 6.57PQMLLLAKK186 pKa = 9.04HH187 pKa = 5.98HH188 pKa = 6.28RR189 pKa = 11.84VIPSRR194 pKa = 11.84QRR196 pKa = 11.84KK197 pKa = 8.29PLGKK201 pKa = 9.48PYY203 pKa = 10.24IKK205 pKa = 10.78VKK207 pKa = 10.4IKK209 pKa = 10.25PPKK212 pKa = 9.87QMLTEE217 pKa = 4.24WFFQQDD223 pKa = 3.38FATQGLFSIHH233 pKa = 6.24AAAANMQWAYY243 pKa = 11.16YY244 pKa = 10.72NPDD247 pKa = 3.24QQSRR251 pKa = 11.84CLTLYY256 pKa = 10.61SLNIKK261 pKa = 9.91FFANTDD267 pKa = 2.75WSATHH272 pKa = 7.08TGTQMYY278 pKa = 9.25FPYY281 pKa = 10.41NAWPHH286 pKa = 5.26TKK288 pKa = 10.37GLTFWTIDD296 pKa = 3.27SKK298 pKa = 11.74GNKK301 pKa = 7.88TPHH304 pKa = 6.43SIKK307 pKa = 9.57PQNYY311 pKa = 7.95NQSVNYY317 pKa = 9.75LQGFFSQYY325 pKa = 10.04VLQAQKK331 pKa = 9.05ITDD334 pKa = 3.69ADD336 pKa = 4.11DD337 pKa = 3.84NVPAKK342 pKa = 10.65YY343 pKa = 9.15EE344 pKa = 4.04QYY346 pKa = 10.2SAAMSTLLIAAFRR359 pKa = 11.84YY360 pKa = 9.99NPEE363 pKa = 3.78EE364 pKa = 3.93DD365 pKa = 3.17TGQGNVVWLTSTLSNTHH382 pKa = 5.59WEE384 pKa = 4.36RR385 pKa = 11.84PQLSDD390 pKa = 3.93LVIVGKK396 pKa = 8.49PLWLSFYY403 pKa = 10.4GFEE406 pKa = 5.12SFIQKK411 pKa = 8.44TKK413 pKa = 10.33HH414 pKa = 5.85DD415 pKa = 4.45KK416 pKa = 10.8GWLLSGFYY424 pKa = 10.21VCKK427 pKa = 10.42CPYY430 pKa = 10.56LKK432 pKa = 9.78TLSATTQTTFAFIDD446 pKa = 3.48LDD448 pKa = 4.39FIQGKK453 pKa = 9.26LPYY456 pKa = 10.1GEE458 pKa = 5.32TITSQKK464 pKa = 9.55KK465 pKa = 9.38ALWYY469 pKa = 9.84PNVEE473 pKa = 4.12NQVNTVTNLVQCGPYY488 pKa = 8.73IPKK491 pKa = 10.15YY492 pKa = 10.36AFRR495 pKa = 11.84DD496 pKa = 3.5EE497 pKa = 4.61STWEE501 pKa = 3.72LTYY504 pKa = 10.74KK505 pKa = 10.69YY506 pKa = 10.18IFYY509 pKa = 9.72FKK511 pKa = 10.09WGGPYY516 pKa = 10.45VSDD519 pKa = 4.94QPVQNPKK526 pKa = 8.61TQGKK530 pKa = 9.31YY531 pKa = 9.39PVPDD535 pKa = 3.63TVSQAIQISNPLKK548 pKa = 8.7QTCKK552 pKa = 10.81AMLRR556 pKa = 11.84DD557 pKa = 2.94WDD559 pKa = 3.74YY560 pKa = 11.54RR561 pKa = 11.84RR562 pKa = 11.84GIITTTALKK571 pKa = 10.57RR572 pKa = 11.84MSEE575 pKa = 4.07HH576 pKa = 6.36LQTDD580 pKa = 3.47SSVQSDD586 pKa = 3.96DD587 pKa = 4.52SEE589 pKa = 4.56TPKK592 pKa = 10.74KK593 pKa = 10.31KK594 pKa = 10.5EE595 pKa = 3.71KK596 pKa = 7.79TTAEE600 pKa = 3.46IPYY603 pKa = 8.36YY604 pKa = 8.52QEE606 pKa = 3.77KK607 pKa = 8.56TEE609 pKa = 4.12EE610 pKa = 4.1MQSCLLSLFEE620 pKa = 5.35EE621 pKa = 5.16STCQEE626 pKa = 4.27PEE628 pKa = 3.85NLQQLILHH636 pKa = 5.81QQQQQQKK643 pKa = 9.04LKK645 pKa = 11.13SNILKK650 pKa = 10.6LLMDD654 pKa = 5.3LKK656 pKa = 10.31HH657 pKa = 5.45QQRR660 pKa = 11.84ILQHH664 pKa = 5.33HH665 pKa = 6.26TGIEE669 pKa = 3.91
MM1 pKa = 7.31PFWWARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.25KK10 pKa = 9.8PWYY13 pKa = 9.88SNWNRR18 pKa = 11.84YY19 pKa = 6.51RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 8.8KK24 pKa = 9.61RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84WPRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84IPYY38 pKa = 7.85GRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 8.72RR49 pKa = 11.84KK50 pKa = 8.81VRR52 pKa = 11.84RR53 pKa = 11.84KK54 pKa = 7.41RR55 pKa = 11.84QRR57 pKa = 11.84IIVKK61 pKa = 8.67QWQPEE66 pKa = 4.43SIVKK70 pKa = 10.5CEE72 pKa = 3.87IKK74 pKa = 10.97GLGCLVAGAQGRR86 pKa = 11.84QPFCYY91 pKa = 9.55TNSKK95 pKa = 10.36YY96 pKa = 10.54EE97 pKa = 3.96YY98 pKa = 8.79PQPKK102 pKa = 10.0APGGGGFGAEE112 pKa = 4.52LFTLQYY118 pKa = 11.06LYY120 pKa = 10.94DD121 pKa = 3.44EE122 pKa = 4.42WVARR126 pKa = 11.84RR127 pKa = 11.84NIWTKK132 pKa = 9.79TNDD135 pKa = 3.36YY136 pKa = 10.38KK137 pKa = 11.52DD138 pKa = 3.57LVRR141 pKa = 11.84FTGATFYY148 pKa = 10.72LYY150 pKa = 10.58KK151 pKa = 10.68HH152 pKa = 6.27PTTDD156 pKa = 5.35FIFNYY161 pKa = 10.01NNQPPFLLEE170 pKa = 3.59KK171 pKa = 10.73DD172 pKa = 4.62FYY174 pKa = 11.67NDD176 pKa = 3.27LHH178 pKa = 6.57PQMLLLAKK186 pKa = 9.04HH187 pKa = 5.98HH188 pKa = 6.28RR189 pKa = 11.84VIPSRR194 pKa = 11.84QRR196 pKa = 11.84KK197 pKa = 8.29PLGKK201 pKa = 9.48PYY203 pKa = 10.24IKK205 pKa = 10.78VKK207 pKa = 10.4IKK209 pKa = 10.25PPKK212 pKa = 9.87QMLTEE217 pKa = 4.24WFFQQDD223 pKa = 3.38FATQGLFSIHH233 pKa = 6.24AAAANMQWAYY243 pKa = 11.16YY244 pKa = 10.72NPDD247 pKa = 3.24QQSRR251 pKa = 11.84CLTLYY256 pKa = 10.61SLNIKK261 pKa = 9.91FFANTDD267 pKa = 2.75WSATHH272 pKa = 7.08TGTQMYY278 pKa = 9.25FPYY281 pKa = 10.41NAWPHH286 pKa = 5.26TKK288 pKa = 10.37GLTFWTIDD296 pKa = 3.27SKK298 pKa = 11.74GNKK301 pKa = 7.88TPHH304 pKa = 6.43SIKK307 pKa = 9.57PQNYY311 pKa = 7.95NQSVNYY317 pKa = 9.75LQGFFSQYY325 pKa = 10.04VLQAQKK331 pKa = 9.05ITDD334 pKa = 3.69ADD336 pKa = 4.11DD337 pKa = 3.84NVPAKK342 pKa = 10.65YY343 pKa = 9.15EE344 pKa = 4.04QYY346 pKa = 10.2SAAMSTLLIAAFRR359 pKa = 11.84YY360 pKa = 9.99NPEE363 pKa = 3.78EE364 pKa = 3.93DD365 pKa = 3.17TGQGNVVWLTSTLSNTHH382 pKa = 5.59WEE384 pKa = 4.36RR385 pKa = 11.84PQLSDD390 pKa = 3.93LVIVGKK396 pKa = 8.49PLWLSFYY403 pKa = 10.4GFEE406 pKa = 5.12SFIQKK411 pKa = 8.44TKK413 pKa = 10.33HH414 pKa = 5.85DD415 pKa = 4.45KK416 pKa = 10.8GWLLSGFYY424 pKa = 10.21VCKK427 pKa = 10.42CPYY430 pKa = 10.56LKK432 pKa = 9.78TLSATTQTTFAFIDD446 pKa = 3.48LDD448 pKa = 4.39FIQGKK453 pKa = 9.26LPYY456 pKa = 10.1GEE458 pKa = 5.32TITSQKK464 pKa = 9.55KK465 pKa = 9.38ALWYY469 pKa = 9.84PNVEE473 pKa = 4.12NQVNTVTNLVQCGPYY488 pKa = 8.73IPKK491 pKa = 10.15YY492 pKa = 10.36AFRR495 pKa = 11.84DD496 pKa = 3.5EE497 pKa = 4.61STWEE501 pKa = 3.72LTYY504 pKa = 10.74KK505 pKa = 10.69YY506 pKa = 10.18IFYY509 pKa = 9.72FKK511 pKa = 10.09WGGPYY516 pKa = 10.45VSDD519 pKa = 4.94QPVQNPKK526 pKa = 8.61TQGKK530 pKa = 9.31YY531 pKa = 9.39PVPDD535 pKa = 3.63TVSQAIQISNPLKK548 pKa = 8.7QTCKK552 pKa = 10.81AMLRR556 pKa = 11.84DD557 pKa = 2.94WDD559 pKa = 3.74YY560 pKa = 11.54RR561 pKa = 11.84RR562 pKa = 11.84GIITTTALKK571 pKa = 10.57RR572 pKa = 11.84MSEE575 pKa = 4.07HH576 pKa = 6.36LQTDD580 pKa = 3.47SSVQSDD586 pKa = 3.96DD587 pKa = 4.52SEE589 pKa = 4.56TPKK592 pKa = 10.74KK593 pKa = 10.31KK594 pKa = 10.5EE595 pKa = 3.71KK596 pKa = 7.79TTAEE600 pKa = 3.46IPYY603 pKa = 8.36YY604 pKa = 8.52QEE606 pKa = 3.77KK607 pKa = 8.56TEE609 pKa = 4.12EE610 pKa = 4.1MQSCLLSLFEE620 pKa = 5.35EE621 pKa = 5.16STCQEE626 pKa = 4.27PEE628 pKa = 3.85NLQQLILHH636 pKa = 5.81QQQQQQKK643 pKa = 9.04LKK645 pKa = 11.13SNILKK650 pKa = 10.6LLMDD654 pKa = 5.3LKK656 pKa = 10.31HH657 pKa = 5.45QQRR660 pKa = 11.84ILQHH664 pKa = 5.33HH665 pKa = 6.26TGIEE669 pKa = 3.91
Molecular weight: 79.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1306 |
122 |
669 |
326.5 |
37.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.662 ± 1.463 | 2.374 ± 0.598 |
4.594 ± 0.662 | 6.126 ± 1.358 |
4.058 ± 0.548 | 5.13 ± 0.426 |
3.292 ± 0.653 | 4.518 ± 0.325 |
8.729 ± 0.736 | 8.116 ± 0.309 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.761 ± 0.181 | 3.752 ± 0.545 |
5.896 ± 0.976 | 6.738 ± 1.181 |
5.666 ± 0.735 | 6.891 ± 1.628 |
6.968 ± 0.409 | 2.527 ± 0.798 |
2.144 ± 0.611 | 4.058 ± 1.268 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
