
Amphibacillus marinus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Amphibacillus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
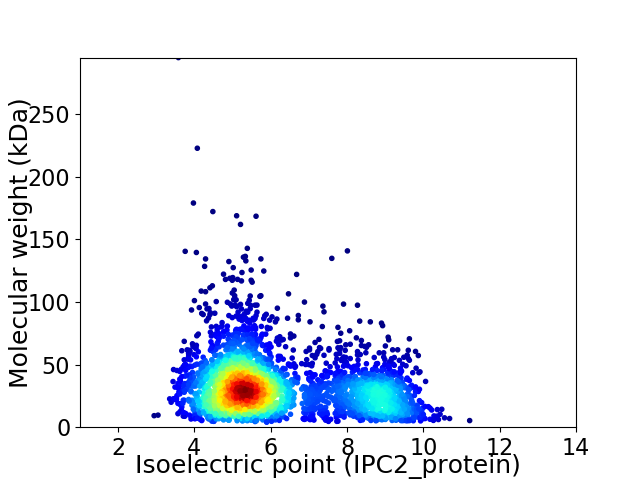
Virtual 2D-PAGE plot for 3270 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8HDV2|A0A1H8HDV2_9BACI Competence protein ComGD OS=Amphibacillus marinus OX=872970 GN=SAMN04488134_101322 PE=4 SV=1
MM1 pKa = 8.3DD2 pKa = 4.72SLRR5 pKa = 11.84KK6 pKa = 9.78RR7 pKa = 11.84GVYY10 pKa = 10.18SLAGLFLLFVLVACGGNDD28 pKa = 3.34GTATNDD34 pKa = 3.04SSGEE38 pKa = 4.02GGEE41 pKa = 4.26SGEE44 pKa = 4.08TTLINEE50 pKa = 4.32ITIMAPTFEE59 pKa = 4.54TTAPPADD66 pKa = 4.24NEE68 pKa = 3.99WEE70 pKa = 4.14QAVEE74 pKa = 4.01AFTGRR79 pKa = 11.84QLTINWVPNVSYY91 pKa = 10.41EE92 pKa = 4.0DD93 pKa = 3.58RR94 pKa = 11.84MNVTLASTDD103 pKa = 3.25IPHH106 pKa = 6.29VMVIQGKK113 pKa = 7.32TPGFLNSAEE122 pKa = 4.66AGAFWEE128 pKa = 4.55LSDD131 pKa = 4.63YY132 pKa = 11.34LDD134 pKa = 4.12DD135 pKa = 5.0YY136 pKa = 11.57EE137 pKa = 5.72YY138 pKa = 11.17LSQYY142 pKa = 11.0NEE144 pKa = 4.77DD145 pKa = 3.48ILRR148 pKa = 11.84NASVNGNVYY157 pKa = 10.44GLYY160 pKa = 9.94RR161 pKa = 11.84GRR163 pKa = 11.84DD164 pKa = 3.33IMRR167 pKa = 11.84STAIVRR173 pKa = 11.84KK174 pKa = 9.83DD175 pKa = 2.91WLDD178 pKa = 3.48NLGLDD183 pKa = 3.76VPEE186 pKa = 5.29TIDD189 pKa = 4.39DD190 pKa = 4.25LYY192 pKa = 11.49EE193 pKa = 3.75VLQAFTHH200 pKa = 7.24DD201 pKa = 4.44DD202 pKa = 3.66PDD204 pKa = 5.15GNGADD209 pKa = 3.97DD210 pKa = 4.09TVGLIIPTWYY220 pKa = 10.68GSLDD224 pKa = 3.52TLSIWFGAPNVWGYY238 pKa = 11.52EE239 pKa = 3.88NGEE242 pKa = 4.23VVPAFTTDD250 pKa = 2.76AYY252 pKa = 9.92KK253 pKa = 10.97ASLEE257 pKa = 4.07FVKK260 pKa = 10.82KK261 pKa = 10.39IVDD264 pKa = 3.65EE265 pKa = 4.45GLVNPDD271 pKa = 3.59FTTLSPDD278 pKa = 3.11DD279 pKa = 3.95WNNAMFNGQGGVIIDD294 pKa = 4.07TYY296 pKa = 10.83SRR298 pKa = 11.84AMQINNLFVDD308 pKa = 4.38EE309 pKa = 4.81TGSTDD314 pKa = 3.56PEE316 pKa = 4.01EE317 pKa = 4.73HH318 pKa = 6.01FVEE321 pKa = 4.05ITGTIRR327 pKa = 11.84SSDD330 pKa = 3.56GEE332 pKa = 4.38EE333 pKa = 3.99YY334 pKa = 10.86GQPTDD339 pKa = 3.94GYY341 pKa = 10.57SGFLAISKK349 pKa = 9.15TSVQTEE355 pKa = 4.38EE356 pKa = 4.26EE357 pKa = 4.13LHH359 pKa = 6.11EE360 pKa = 4.21VLAFLDD366 pKa = 3.86QLSSADD372 pKa = 3.99GLNLLNHH379 pKa = 7.24GIEE382 pKa = 4.29GVNYY386 pKa = 9.44EE387 pKa = 4.73LDD389 pKa = 3.78DD390 pKa = 4.29EE391 pKa = 5.67GYY393 pKa = 10.54LIPLEE398 pKa = 4.34TPEE401 pKa = 4.07AAALKK406 pKa = 9.09PYY408 pKa = 10.93EE409 pKa = 4.26MGQISTYY416 pKa = 10.5GEE418 pKa = 4.2GMHH421 pKa = 6.88TMRR424 pKa = 11.84QDD426 pKa = 2.87GALPQKK432 pKa = 10.2RR433 pKa = 11.84YY434 pKa = 9.86QLMEE438 pKa = 4.99DD439 pKa = 3.81NEE441 pKa = 4.01AHH443 pKa = 6.11AVYY446 pKa = 10.35NAAAPLVSEE455 pKa = 4.88VYY457 pKa = 7.0TTRR460 pKa = 11.84GTQLDD465 pKa = 4.84DD466 pKa = 4.55IIADD470 pKa = 3.51ARR472 pKa = 11.84IQYY475 pKa = 9.35VAGHH479 pKa = 6.65IDD481 pKa = 3.38EE482 pKa = 5.69AGWDD486 pKa = 3.62SAIDD490 pKa = 3.34LWYY493 pKa = 10.97SSGGQEE499 pKa = 6.15LIDD502 pKa = 4.4EE503 pKa = 4.65LNEE506 pKa = 5.01LYY508 pKa = 10.52TEE510 pKa = 4.38MNN512 pKa = 3.48
MM1 pKa = 8.3DD2 pKa = 4.72SLRR5 pKa = 11.84KK6 pKa = 9.78RR7 pKa = 11.84GVYY10 pKa = 10.18SLAGLFLLFVLVACGGNDD28 pKa = 3.34GTATNDD34 pKa = 3.04SSGEE38 pKa = 4.02GGEE41 pKa = 4.26SGEE44 pKa = 4.08TTLINEE50 pKa = 4.32ITIMAPTFEE59 pKa = 4.54TTAPPADD66 pKa = 4.24NEE68 pKa = 3.99WEE70 pKa = 4.14QAVEE74 pKa = 4.01AFTGRR79 pKa = 11.84QLTINWVPNVSYY91 pKa = 10.41EE92 pKa = 4.0DD93 pKa = 3.58RR94 pKa = 11.84MNVTLASTDD103 pKa = 3.25IPHH106 pKa = 6.29VMVIQGKK113 pKa = 7.32TPGFLNSAEE122 pKa = 4.66AGAFWEE128 pKa = 4.55LSDD131 pKa = 4.63YY132 pKa = 11.34LDD134 pKa = 4.12DD135 pKa = 5.0YY136 pKa = 11.57EE137 pKa = 5.72YY138 pKa = 11.17LSQYY142 pKa = 11.0NEE144 pKa = 4.77DD145 pKa = 3.48ILRR148 pKa = 11.84NASVNGNVYY157 pKa = 10.44GLYY160 pKa = 9.94RR161 pKa = 11.84GRR163 pKa = 11.84DD164 pKa = 3.33IMRR167 pKa = 11.84STAIVRR173 pKa = 11.84KK174 pKa = 9.83DD175 pKa = 2.91WLDD178 pKa = 3.48NLGLDD183 pKa = 3.76VPEE186 pKa = 5.29TIDD189 pKa = 4.39DD190 pKa = 4.25LYY192 pKa = 11.49EE193 pKa = 3.75VLQAFTHH200 pKa = 7.24DD201 pKa = 4.44DD202 pKa = 3.66PDD204 pKa = 5.15GNGADD209 pKa = 3.97DD210 pKa = 4.09TVGLIIPTWYY220 pKa = 10.68GSLDD224 pKa = 3.52TLSIWFGAPNVWGYY238 pKa = 11.52EE239 pKa = 3.88NGEE242 pKa = 4.23VVPAFTTDD250 pKa = 2.76AYY252 pKa = 9.92KK253 pKa = 10.97ASLEE257 pKa = 4.07FVKK260 pKa = 10.82KK261 pKa = 10.39IVDD264 pKa = 3.65EE265 pKa = 4.45GLVNPDD271 pKa = 3.59FTTLSPDD278 pKa = 3.11DD279 pKa = 3.95WNNAMFNGQGGVIIDD294 pKa = 4.07TYY296 pKa = 10.83SRR298 pKa = 11.84AMQINNLFVDD308 pKa = 4.38EE309 pKa = 4.81TGSTDD314 pKa = 3.56PEE316 pKa = 4.01EE317 pKa = 4.73HH318 pKa = 6.01FVEE321 pKa = 4.05ITGTIRR327 pKa = 11.84SSDD330 pKa = 3.56GEE332 pKa = 4.38EE333 pKa = 3.99YY334 pKa = 10.86GQPTDD339 pKa = 3.94GYY341 pKa = 10.57SGFLAISKK349 pKa = 9.15TSVQTEE355 pKa = 4.38EE356 pKa = 4.26EE357 pKa = 4.13LHH359 pKa = 6.11EE360 pKa = 4.21VLAFLDD366 pKa = 3.86QLSSADD372 pKa = 3.99GLNLLNHH379 pKa = 7.24GIEE382 pKa = 4.29GVNYY386 pKa = 9.44EE387 pKa = 4.73LDD389 pKa = 3.78DD390 pKa = 4.29EE391 pKa = 5.67GYY393 pKa = 10.54LIPLEE398 pKa = 4.34TPEE401 pKa = 4.07AAALKK406 pKa = 9.09PYY408 pKa = 10.93EE409 pKa = 4.26MGQISTYY416 pKa = 10.5GEE418 pKa = 4.2GMHH421 pKa = 6.88TMRR424 pKa = 11.84QDD426 pKa = 2.87GALPQKK432 pKa = 10.2RR433 pKa = 11.84YY434 pKa = 9.86QLMEE438 pKa = 4.99DD439 pKa = 3.81NEE441 pKa = 4.01AHH443 pKa = 6.11AVYY446 pKa = 10.35NAAAPLVSEE455 pKa = 4.88VYY457 pKa = 7.0TTRR460 pKa = 11.84GTQLDD465 pKa = 4.84DD466 pKa = 4.55IIADD470 pKa = 3.51ARR472 pKa = 11.84IQYY475 pKa = 9.35VAGHH479 pKa = 6.65IDD481 pKa = 3.38EE482 pKa = 5.69AGWDD486 pKa = 3.62SAIDD490 pKa = 3.34LWYY493 pKa = 10.97SSGGQEE499 pKa = 6.15LIDD502 pKa = 4.4EE503 pKa = 4.65LNEE506 pKa = 5.01LYY508 pKa = 10.52TEE510 pKa = 4.38MNN512 pKa = 3.48
Molecular weight: 56.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8PIX5|A0A1H8PIX5_9BACI 50S ribosomal protein L9 OS=Amphibacillus marinus OX=872970 GN=rplI PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84HH29 pKa = 3.69VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.73KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84HH29 pKa = 3.69VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.73KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
988883 |
39 |
2731 |
302.4 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.444 ± 0.047 | 0.67 ± 0.012 |
5.415 ± 0.036 | 6.978 ± 0.045 |
4.532 ± 0.034 | 6.445 ± 0.039 |
2.19 ± 0.02 | 8.069 ± 0.035 |
5.798 ± 0.042 | 10.182 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.475 ± 0.019 | 4.624 ± 0.029 |
3.421 ± 0.024 | 4.711 ± 0.033 |
4.13 ± 0.028 | 5.813 ± 0.031 |
5.579 ± 0.033 | 6.825 ± 0.032 |
0.988 ± 0.014 | 3.709 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
