
Beihai barnacle virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
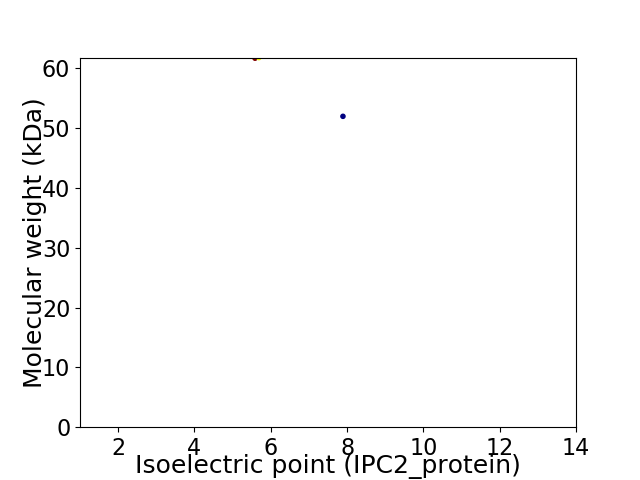
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
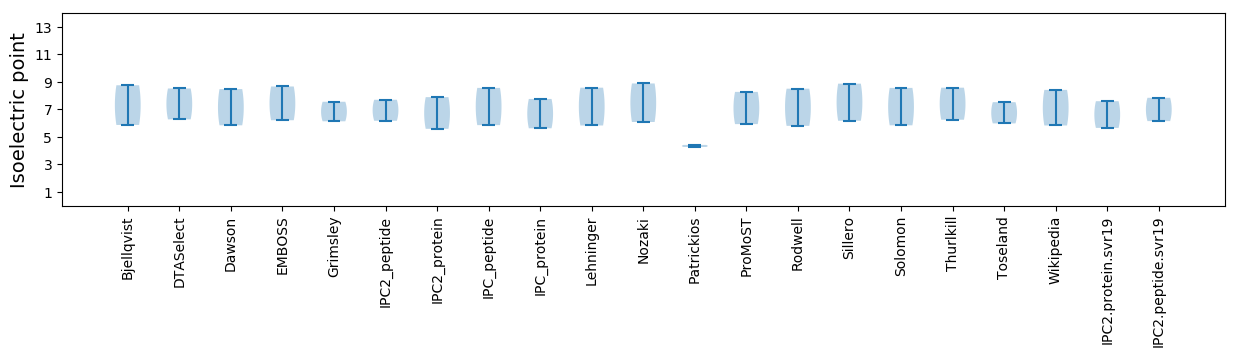
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLA3|A0A1L3KLA3_9VIRU RdRp OS=Beihai barnacle virus 12 OX=1922356 PE=4 SV=1
MM1 pKa = 7.74LSLQPGEE8 pKa = 4.58SFRR11 pKa = 11.84QDD13 pKa = 2.87FRR15 pKa = 11.84NPGRR19 pKa = 11.84DD20 pKa = 3.27KK21 pKa = 11.29LATEE25 pKa = 4.61ALNHH29 pKa = 6.35AFGSDD34 pKa = 3.22RR35 pKa = 11.84VDD37 pKa = 3.96KK38 pKa = 10.98ILATLHH44 pKa = 6.26RR45 pKa = 11.84AEE47 pKa = 4.83KK48 pKa = 10.89SLDD51 pKa = 3.48EE52 pKa = 5.07LIADD56 pKa = 3.37VMLFDD61 pKa = 4.74RR62 pKa = 11.84EE63 pKa = 4.34RR64 pKa = 11.84VGPAARR70 pKa = 11.84DD71 pKa = 3.56DD72 pKa = 4.08PVYY75 pKa = 10.73QLALEE80 pKa = 4.31SVRR83 pKa = 11.84QEE85 pKa = 4.49FIPKK89 pKa = 6.21EE90 pKa = 4.26TKK92 pKa = 10.4VIPLTLGGVEE102 pKa = 4.48NRR104 pKa = 11.84PDD106 pKa = 3.37LPRR109 pKa = 11.84NKK111 pKa = 10.42SPGLPLNEE119 pKa = 3.94HH120 pKa = 6.79FKK122 pKa = 10.84TKK124 pKa = 10.81GEE126 pKa = 4.05ALSDD130 pKa = 3.94PGVHH134 pKa = 7.34ADD136 pKa = 3.61IHH138 pKa = 6.67KK139 pKa = 9.99KK140 pKa = 8.22WALIGKK146 pKa = 8.73GYY148 pKa = 9.19KK149 pKa = 9.48QKK151 pKa = 10.83LHH153 pKa = 7.24DD154 pKa = 3.84SALFQRR160 pKa = 11.84AQICSTEE167 pKa = 4.03KK168 pKa = 10.57NKK170 pKa = 9.4IRR172 pKa = 11.84AVWGYY177 pKa = 8.1PLEE180 pKa = 4.31VFMEE184 pKa = 4.67EE185 pKa = 4.04ARR187 pKa = 11.84FFYY190 pKa = 10.06PYY192 pKa = 9.34MDD194 pKa = 5.19YY195 pKa = 10.81ILSCAQNLPIGYY207 pKa = 8.24QAEE210 pKa = 4.34MATGGMLYY218 pKa = 9.86INDD221 pKa = 4.6MIRR224 pKa = 11.84SHH226 pKa = 6.5PHH228 pKa = 4.43ATFAICDD235 pKa = 3.35WSRR238 pKa = 11.84FDD240 pKa = 3.4KK241 pKa = 10.52TVPAWLIRR249 pKa = 11.84DD250 pKa = 4.24AFQIILDD257 pKa = 4.02SLDD260 pKa = 3.35LSKK263 pKa = 11.44VQGSDD268 pKa = 3.08GAIWNVRR275 pKa = 11.84EE276 pKa = 3.97EE277 pKa = 4.12DD278 pKa = 3.24SLRR281 pKa = 11.84RR282 pKa = 11.84FKK284 pKa = 10.9RR285 pKa = 11.84VVSYY289 pKa = 8.48FINTPIRR296 pKa = 11.84SPDD299 pKa = 2.78GSRR302 pKa = 11.84FRR304 pKa = 11.84KK305 pKa = 9.95RR306 pKa = 11.84GGVPSGSAFTNIIDD320 pKa = 4.27SIVNAIVTRR329 pKa = 11.84YY330 pKa = 9.11LAYY333 pKa = 9.8HH334 pKa = 6.96CSGALPSADD343 pKa = 3.8IYY345 pKa = 11.42LGDD348 pKa = 4.91DD349 pKa = 3.55SVCVVNGVVNLQDD362 pKa = 3.19WADD365 pKa = 3.69LARR368 pKa = 11.84EE369 pKa = 4.06KK370 pKa = 10.94FSMEE374 pKa = 3.96LNVEE378 pKa = 3.95KK379 pKa = 10.7SYY381 pKa = 10.1VTTNPCNVHH390 pKa = 5.57FLGYY394 pKa = 10.49FNLQGLPIKK403 pKa = 10.74GQDD406 pKa = 3.01FAMASFLYY414 pKa = 9.78PEE416 pKa = 4.89RR417 pKa = 11.84KK418 pKa = 9.58SLSPTITAARR428 pKa = 11.84ALGQMWSTMNPYY440 pKa = 9.97AASNWHH446 pKa = 6.73DD447 pKa = 3.51VVTYY451 pKa = 10.92IMDD454 pKa = 3.96TQDD457 pKa = 2.93VTLSQIQEE465 pKa = 4.15HH466 pKa = 6.47LRR468 pKa = 11.84SQPEE472 pKa = 3.66AFRR475 pKa = 11.84YY476 pKa = 9.53LRR478 pKa = 11.84MMGLDD483 pKa = 3.47VATLGLPRR491 pKa = 11.84KK492 pKa = 9.85VDD494 pKa = 3.54GLVLEE499 pKa = 4.3VQPRR503 pKa = 11.84SVSRR507 pKa = 11.84KK508 pKa = 7.21IYY510 pKa = 10.41KK511 pKa = 9.9VVTHH515 pKa = 7.26DD516 pKa = 3.42VTKK519 pKa = 10.89LMSLAIDD526 pKa = 4.6RR527 pKa = 11.84IEE529 pKa = 4.44TDD531 pKa = 3.38LQEE534 pKa = 4.31QDD536 pKa = 4.3DD537 pKa = 4.42VPEE540 pKa = 4.58EE541 pKa = 4.11EE542 pKa = 5.4DD543 pKa = 4.47DD544 pKa = 3.71NN545 pKa = 4.67
MM1 pKa = 7.74LSLQPGEE8 pKa = 4.58SFRR11 pKa = 11.84QDD13 pKa = 2.87FRR15 pKa = 11.84NPGRR19 pKa = 11.84DD20 pKa = 3.27KK21 pKa = 11.29LATEE25 pKa = 4.61ALNHH29 pKa = 6.35AFGSDD34 pKa = 3.22RR35 pKa = 11.84VDD37 pKa = 3.96KK38 pKa = 10.98ILATLHH44 pKa = 6.26RR45 pKa = 11.84AEE47 pKa = 4.83KK48 pKa = 10.89SLDD51 pKa = 3.48EE52 pKa = 5.07LIADD56 pKa = 3.37VMLFDD61 pKa = 4.74RR62 pKa = 11.84EE63 pKa = 4.34RR64 pKa = 11.84VGPAARR70 pKa = 11.84DD71 pKa = 3.56DD72 pKa = 4.08PVYY75 pKa = 10.73QLALEE80 pKa = 4.31SVRR83 pKa = 11.84QEE85 pKa = 4.49FIPKK89 pKa = 6.21EE90 pKa = 4.26TKK92 pKa = 10.4VIPLTLGGVEE102 pKa = 4.48NRR104 pKa = 11.84PDD106 pKa = 3.37LPRR109 pKa = 11.84NKK111 pKa = 10.42SPGLPLNEE119 pKa = 3.94HH120 pKa = 6.79FKK122 pKa = 10.84TKK124 pKa = 10.81GEE126 pKa = 4.05ALSDD130 pKa = 3.94PGVHH134 pKa = 7.34ADD136 pKa = 3.61IHH138 pKa = 6.67KK139 pKa = 9.99KK140 pKa = 8.22WALIGKK146 pKa = 8.73GYY148 pKa = 9.19KK149 pKa = 9.48QKK151 pKa = 10.83LHH153 pKa = 7.24DD154 pKa = 3.84SALFQRR160 pKa = 11.84AQICSTEE167 pKa = 4.03KK168 pKa = 10.57NKK170 pKa = 9.4IRR172 pKa = 11.84AVWGYY177 pKa = 8.1PLEE180 pKa = 4.31VFMEE184 pKa = 4.67EE185 pKa = 4.04ARR187 pKa = 11.84FFYY190 pKa = 10.06PYY192 pKa = 9.34MDD194 pKa = 5.19YY195 pKa = 10.81ILSCAQNLPIGYY207 pKa = 8.24QAEE210 pKa = 4.34MATGGMLYY218 pKa = 9.86INDD221 pKa = 4.6MIRR224 pKa = 11.84SHH226 pKa = 6.5PHH228 pKa = 4.43ATFAICDD235 pKa = 3.35WSRR238 pKa = 11.84FDD240 pKa = 3.4KK241 pKa = 10.52TVPAWLIRR249 pKa = 11.84DD250 pKa = 4.24AFQIILDD257 pKa = 4.02SLDD260 pKa = 3.35LSKK263 pKa = 11.44VQGSDD268 pKa = 3.08GAIWNVRR275 pKa = 11.84EE276 pKa = 3.97EE277 pKa = 4.12DD278 pKa = 3.24SLRR281 pKa = 11.84RR282 pKa = 11.84FKK284 pKa = 10.9RR285 pKa = 11.84VVSYY289 pKa = 8.48FINTPIRR296 pKa = 11.84SPDD299 pKa = 2.78GSRR302 pKa = 11.84FRR304 pKa = 11.84KK305 pKa = 9.95RR306 pKa = 11.84GGVPSGSAFTNIIDD320 pKa = 4.27SIVNAIVTRR329 pKa = 11.84YY330 pKa = 9.11LAYY333 pKa = 9.8HH334 pKa = 6.96CSGALPSADD343 pKa = 3.8IYY345 pKa = 11.42LGDD348 pKa = 4.91DD349 pKa = 3.55SVCVVNGVVNLQDD362 pKa = 3.19WADD365 pKa = 3.69LARR368 pKa = 11.84EE369 pKa = 4.06KK370 pKa = 10.94FSMEE374 pKa = 3.96LNVEE378 pKa = 3.95KK379 pKa = 10.7SYY381 pKa = 10.1VTTNPCNVHH390 pKa = 5.57FLGYY394 pKa = 10.49FNLQGLPIKK403 pKa = 10.74GQDD406 pKa = 3.01FAMASFLYY414 pKa = 9.78PEE416 pKa = 4.89RR417 pKa = 11.84KK418 pKa = 9.58SLSPTITAARR428 pKa = 11.84ALGQMWSTMNPYY440 pKa = 9.97AASNWHH446 pKa = 6.73DD447 pKa = 3.51VVTYY451 pKa = 10.92IMDD454 pKa = 3.96TQDD457 pKa = 2.93VTLSQIQEE465 pKa = 4.15HH466 pKa = 6.47LRR468 pKa = 11.84SQPEE472 pKa = 3.66AFRR475 pKa = 11.84YY476 pKa = 9.53LRR478 pKa = 11.84MMGLDD483 pKa = 3.47VATLGLPRR491 pKa = 11.84KK492 pKa = 9.85VDD494 pKa = 3.54GLVLEE499 pKa = 4.3VQPRR503 pKa = 11.84SVSRR507 pKa = 11.84KK508 pKa = 7.21IYY510 pKa = 10.41KK511 pKa = 9.9VVTHH515 pKa = 7.26DD516 pKa = 3.42VTKK519 pKa = 10.89LMSLAIDD526 pKa = 4.6RR527 pKa = 11.84IEE529 pKa = 4.44TDD531 pKa = 3.38LQEE534 pKa = 4.31QDD536 pKa = 4.3DD537 pKa = 4.42VPEE540 pKa = 4.58EE541 pKa = 4.11EE542 pKa = 5.4DD543 pKa = 4.47DD544 pKa = 3.71NN545 pKa = 4.67
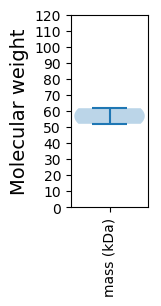
Molecular weight: 61.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLA3|A0A1L3KLA3_9VIRU RdRp OS=Beihai barnacle virus 12 OX=1922356 PE=4 SV=1
MM1 pKa = 7.88SSRR4 pKa = 11.84QTQEE8 pKa = 4.82DD9 pKa = 3.98NQPDD13 pKa = 4.01EE14 pKa = 4.55PVPNPTEE21 pKa = 3.59QKK23 pKa = 9.13TRR25 pKa = 11.84VFSNSGRR32 pKa = 11.84RR33 pKa = 11.84GKK35 pKa = 9.66PKK37 pKa = 9.72PQSKK41 pKa = 10.17AGKK44 pKa = 8.37PKK46 pKa = 10.41VSAGPKK52 pKa = 9.34APPPAGPPKK61 pKa = 9.46PQHH64 pKa = 6.4LSGKK68 pKa = 8.14GTSLANDD75 pKa = 4.49PLWQEE80 pKa = 4.09AFVLEE85 pKa = 4.2GDD87 pKa = 3.38KK88 pKa = 11.28GYY90 pKa = 10.95VQVPTKK96 pKa = 10.75RR97 pKa = 11.84EE98 pKa = 3.92FQVSAEE104 pKa = 4.04GLHH107 pKa = 7.01DD108 pKa = 3.73IASAVYY114 pKa = 9.44TEE116 pKa = 4.07MSMVDD121 pKa = 2.54GHH123 pKa = 5.85FRR125 pKa = 11.84KK126 pKa = 8.58TIPEE130 pKa = 4.32SAFHH134 pKa = 6.76YY135 pKa = 7.64YY136 pKa = 10.27TIVLLHH142 pKa = 6.64ARR144 pKa = 11.84LAEE147 pKa = 4.12LGKK150 pKa = 10.24RR151 pKa = 11.84HH152 pKa = 5.96PTDD155 pKa = 2.78TTYY158 pKa = 11.52EE159 pKa = 3.9DD160 pKa = 3.68FRR162 pKa = 11.84FVEE165 pKa = 4.29LMKK168 pKa = 10.57EE169 pKa = 4.03YY170 pKa = 10.97GPVPEE175 pKa = 5.37PIEE178 pKa = 4.14AYY180 pKa = 9.82FRR182 pKa = 11.84SIGDD186 pKa = 3.79LPDD189 pKa = 3.38VAGVEE194 pKa = 4.58YY195 pKa = 10.69KK196 pKa = 10.82LKK198 pKa = 10.87LPPKK202 pKa = 9.36PLADD206 pKa = 3.17RR207 pKa = 11.84HH208 pKa = 5.7GPSTGFHH215 pKa = 5.85RR216 pKa = 11.84RR217 pKa = 11.84VSAEE221 pKa = 3.73THH223 pKa = 5.12NTWEE227 pKa = 4.17QVPAPGVAAMRR238 pKa = 11.84MCQDD242 pKa = 2.94MAYY245 pKa = 7.7TQHH248 pKa = 7.28PNRR251 pKa = 11.84DD252 pKa = 3.28IGPIWDD258 pKa = 4.45LPAEE262 pKa = 4.05LRR264 pKa = 11.84PVPEE268 pKa = 5.2HH269 pKa = 5.78GHH271 pKa = 5.06TPGRR275 pKa = 11.84PTGNLLGWQPATLLSNEE292 pKa = 3.76QRR294 pKa = 11.84TSLSQCGVTPTEE306 pKa = 3.96ASTAGTYY313 pKa = 10.01AVNLQVLGMVSSALARR329 pKa = 11.84ATSVYY334 pKa = 9.61RR335 pKa = 11.84CASAAVHH342 pKa = 6.06NSANGSYY349 pKa = 10.48AQATFCEE356 pKa = 4.7VEE358 pKa = 4.37DD359 pKa = 4.34SDD361 pKa = 6.1LGFQRR366 pKa = 11.84RR367 pKa = 11.84QQYY370 pKa = 8.83TSAGLRR376 pKa = 11.84SCTAFQADD384 pKa = 3.36ARR386 pKa = 11.84ISVASRR392 pKa = 11.84NCCFRR397 pKa = 11.84VKK399 pKa = 10.38RR400 pKa = 11.84KK401 pKa = 8.26TGADD405 pKa = 3.28GTNRR409 pKa = 11.84WACYY413 pKa = 10.19DD414 pKa = 3.46YY415 pKa = 11.15DD416 pKa = 3.77QYY418 pKa = 11.18RR419 pKa = 11.84TVPPSWTEE427 pKa = 3.61NMNATFNAGEE437 pKa = 4.2ASAEE441 pKa = 3.99LNITKK446 pKa = 10.36LATAYY451 pKa = 10.33ADD453 pKa = 3.32KK454 pKa = 11.12SRR456 pKa = 11.84TLRR459 pKa = 11.84PWARR463 pKa = 11.84APVINKK469 pKa = 9.39SRR471 pKa = 11.84DD472 pKa = 3.13
MM1 pKa = 7.88SSRR4 pKa = 11.84QTQEE8 pKa = 4.82DD9 pKa = 3.98NQPDD13 pKa = 4.01EE14 pKa = 4.55PVPNPTEE21 pKa = 3.59QKK23 pKa = 9.13TRR25 pKa = 11.84VFSNSGRR32 pKa = 11.84RR33 pKa = 11.84GKK35 pKa = 9.66PKK37 pKa = 9.72PQSKK41 pKa = 10.17AGKK44 pKa = 8.37PKK46 pKa = 10.41VSAGPKK52 pKa = 9.34APPPAGPPKK61 pKa = 9.46PQHH64 pKa = 6.4LSGKK68 pKa = 8.14GTSLANDD75 pKa = 4.49PLWQEE80 pKa = 4.09AFVLEE85 pKa = 4.2GDD87 pKa = 3.38KK88 pKa = 11.28GYY90 pKa = 10.95VQVPTKK96 pKa = 10.75RR97 pKa = 11.84EE98 pKa = 3.92FQVSAEE104 pKa = 4.04GLHH107 pKa = 7.01DD108 pKa = 3.73IASAVYY114 pKa = 9.44TEE116 pKa = 4.07MSMVDD121 pKa = 2.54GHH123 pKa = 5.85FRR125 pKa = 11.84KK126 pKa = 8.58TIPEE130 pKa = 4.32SAFHH134 pKa = 6.76YY135 pKa = 7.64YY136 pKa = 10.27TIVLLHH142 pKa = 6.64ARR144 pKa = 11.84LAEE147 pKa = 4.12LGKK150 pKa = 10.24RR151 pKa = 11.84HH152 pKa = 5.96PTDD155 pKa = 2.78TTYY158 pKa = 11.52EE159 pKa = 3.9DD160 pKa = 3.68FRR162 pKa = 11.84FVEE165 pKa = 4.29LMKK168 pKa = 10.57EE169 pKa = 4.03YY170 pKa = 10.97GPVPEE175 pKa = 5.37PIEE178 pKa = 4.14AYY180 pKa = 9.82FRR182 pKa = 11.84SIGDD186 pKa = 3.79LPDD189 pKa = 3.38VAGVEE194 pKa = 4.58YY195 pKa = 10.69KK196 pKa = 10.82LKK198 pKa = 10.87LPPKK202 pKa = 9.36PLADD206 pKa = 3.17RR207 pKa = 11.84HH208 pKa = 5.7GPSTGFHH215 pKa = 5.85RR216 pKa = 11.84RR217 pKa = 11.84VSAEE221 pKa = 3.73THH223 pKa = 5.12NTWEE227 pKa = 4.17QVPAPGVAAMRR238 pKa = 11.84MCQDD242 pKa = 2.94MAYY245 pKa = 7.7TQHH248 pKa = 7.28PNRR251 pKa = 11.84DD252 pKa = 3.28IGPIWDD258 pKa = 4.45LPAEE262 pKa = 4.05LRR264 pKa = 11.84PVPEE268 pKa = 5.2HH269 pKa = 5.78GHH271 pKa = 5.06TPGRR275 pKa = 11.84PTGNLLGWQPATLLSNEE292 pKa = 3.76QRR294 pKa = 11.84TSLSQCGVTPTEE306 pKa = 3.96ASTAGTYY313 pKa = 10.01AVNLQVLGMVSSALARR329 pKa = 11.84ATSVYY334 pKa = 9.61RR335 pKa = 11.84CASAAVHH342 pKa = 6.06NSANGSYY349 pKa = 10.48AQATFCEE356 pKa = 4.7VEE358 pKa = 4.37DD359 pKa = 4.34SDD361 pKa = 6.1LGFQRR366 pKa = 11.84RR367 pKa = 11.84QQYY370 pKa = 8.83TSAGLRR376 pKa = 11.84SCTAFQADD384 pKa = 3.36ARR386 pKa = 11.84ISVASRR392 pKa = 11.84NCCFRR397 pKa = 11.84VKK399 pKa = 10.38RR400 pKa = 11.84KK401 pKa = 8.26TGADD405 pKa = 3.28GTNRR409 pKa = 11.84WACYY413 pKa = 10.19DD414 pKa = 3.46YY415 pKa = 11.15DD416 pKa = 3.77QYY418 pKa = 11.18RR419 pKa = 11.84TVPPSWTEE427 pKa = 3.61NMNATFNAGEE437 pKa = 4.2ASAEE441 pKa = 3.99LNITKK446 pKa = 10.36LATAYY451 pKa = 10.33ADD453 pKa = 3.32KK454 pKa = 11.12SRR456 pKa = 11.84TLRR459 pKa = 11.84PWARR463 pKa = 11.84APVINKK469 pKa = 9.39SRR471 pKa = 11.84DD472 pKa = 3.13
Molecular weight: 51.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1017 |
472 |
545 |
508.5 |
56.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.948 ± 0.998 | 1.377 ± 0.193 |
6.293 ± 0.99 | 5.605 ± 0.07 |
3.638 ± 0.408 | 6.195 ± 0.355 |
2.557 ± 0.12 | 4.031 ± 1.161 |
4.818 ± 0.095 | 7.965 ± 1.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.275 | 3.835 ± 0.013 |
6.883 ± 1.094 | 4.326 ± 0.203 |
6.785 ± 0.126 | 6.981 ± 0.135 |
5.801 ± 0.979 | 6.588 ± 0.269 |
1.475 ± 0.005 | 3.54 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
