
Clostridium sp. CAG:169
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
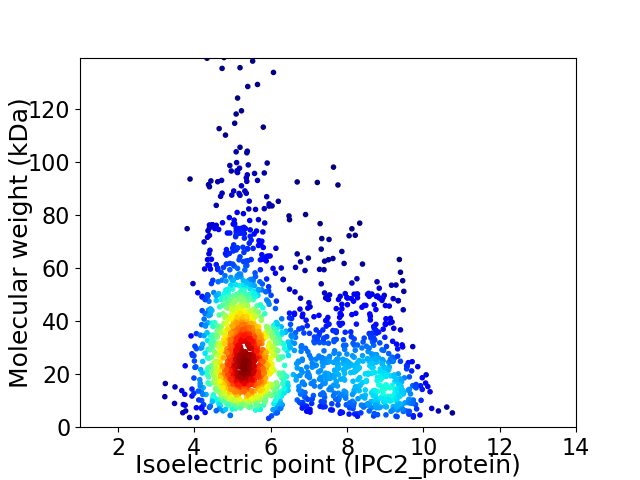
Virtual 2D-PAGE plot for 1970 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6BSZ5|R6BSZ5_9CLOT HNH endonuclease domain protein OS=Clostridium sp. CAG:169 OX=1262778 GN=BN513_01937 PE=4 SV=1
MM1 pKa = 7.64TYY3 pKa = 11.23DD4 pKa = 4.44DD5 pKa = 5.22VITMLEE11 pKa = 4.05EE12 pKa = 4.46AGLPFAYY19 pKa = 10.25DD20 pKa = 3.46HH21 pKa = 6.26FAEE24 pKa = 5.39GEE26 pKa = 4.4SPDD29 pKa = 3.72PPFLVFLYY37 pKa = 9.99PGSDD41 pKa = 2.89NMFADD46 pKa = 3.44DD47 pKa = 4.24TVFKK51 pKa = 10.8KK52 pKa = 10.13IDD54 pKa = 3.47EE55 pKa = 4.6LNIEE59 pKa = 5.05LYY61 pKa = 10.57TDD63 pKa = 3.51VKK65 pKa = 11.09DD66 pKa = 4.63PEE68 pKa = 5.58AEE70 pKa = 4.17TQIEE74 pKa = 4.34DD75 pKa = 3.54TLIAHH80 pKa = 7.29DD81 pKa = 4.46LPYY84 pKa = 10.63EE85 pKa = 4.15KK86 pKa = 10.71SEE88 pKa = 3.77VWIEE92 pKa = 4.0SEE94 pKa = 3.82KK95 pKa = 10.87LYY97 pKa = 10.94EE98 pKa = 4.19VLYY101 pKa = 8.01QTQIIGGG108 pKa = 3.81
MM1 pKa = 7.64TYY3 pKa = 11.23DD4 pKa = 4.44DD5 pKa = 5.22VITMLEE11 pKa = 4.05EE12 pKa = 4.46AGLPFAYY19 pKa = 10.25DD20 pKa = 3.46HH21 pKa = 6.26FAEE24 pKa = 5.39GEE26 pKa = 4.4SPDD29 pKa = 3.72PPFLVFLYY37 pKa = 9.99PGSDD41 pKa = 2.89NMFADD46 pKa = 3.44DD47 pKa = 4.24TVFKK51 pKa = 10.8KK52 pKa = 10.13IDD54 pKa = 3.47EE55 pKa = 4.6LNIEE59 pKa = 5.05LYY61 pKa = 10.57TDD63 pKa = 3.51VKK65 pKa = 11.09DD66 pKa = 4.63PEE68 pKa = 5.58AEE70 pKa = 4.17TQIEE74 pKa = 4.34DD75 pKa = 3.54TLIAHH80 pKa = 7.29DD81 pKa = 4.46LPYY84 pKa = 10.63EE85 pKa = 4.15KK86 pKa = 10.71SEE88 pKa = 3.77VWIEE92 pKa = 4.0SEE94 pKa = 3.82KK95 pKa = 10.87LYY97 pKa = 10.94EE98 pKa = 4.19VLYY101 pKa = 8.01QTQIIGGG108 pKa = 3.81
Molecular weight: 12.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6BC20|R6BC20_9CLOT ABC transmembrane type-1 domain-containing protein OS=Clostridium sp. CAG:169 OX=1262778 GN=BN513_00188 PE=3 SV=1
MM1 pKa = 7.32QAVCCLFVLVGGILPVAQFVHH22 pKa = 6.46PQLDD26 pKa = 3.78HH27 pKa = 7.25LSALHH32 pKa = 6.45HH33 pKa = 5.95RR34 pKa = 11.84RR35 pKa = 11.84CVKK38 pKa = 10.11ILAAHH43 pKa = 6.52GKK45 pKa = 7.64VCRR48 pKa = 11.84IKK50 pKa = 10.44LQRR53 pKa = 11.84VVYY56 pKa = 8.92LVPAQANGSDD66 pKa = 4.42DD67 pKa = 2.99IRR69 pKa = 11.84CRR71 pKa = 11.84VRR73 pKa = 11.84LRR75 pKa = 11.84EE76 pKa = 4.33HH77 pKa = 6.68ILDD80 pKa = 4.48FKK82 pKa = 11.41AGVDD86 pKa = 3.55VPLRR90 pKa = 11.84HH91 pKa = 6.58IMLLHH96 pKa = 6.79LSDD99 pKa = 4.06VLLGQQLVCFSLSHH113 pKa = 6.31NLHH116 pKa = 5.94NFKK119 pKa = 10.22GHH121 pKa = 6.35RR122 pKa = 11.84RR123 pKa = 11.84VQPVVNQIQHH133 pKa = 6.58DD134 pKa = 4.33AVTGTDD140 pKa = 3.18NLADD144 pKa = 3.59VAGAAFDD151 pKa = 3.7EE152 pKa = 4.71VLRR155 pKa = 11.84IAQPYY160 pKa = 8.51VGAVGQTGNLQQIRR174 pKa = 11.84KK175 pKa = 8.73ILRR178 pKa = 11.84LTIQQHH184 pKa = 5.93LPDD187 pKa = 4.1KK188 pKa = 10.69RR189 pKa = 11.84RR190 pKa = 11.84AHH192 pKa = 6.27LRR194 pKa = 11.84DD195 pKa = 3.21GKK197 pKa = 10.96GAGFAVDD204 pKa = 4.88LLRR207 pKa = 11.84CHH209 pKa = 5.83TQHH212 pKa = 6.86LRR214 pKa = 11.84RR215 pKa = 11.84GEE217 pKa = 4.14QAVHH221 pKa = 6.07LLVRR225 pKa = 11.84HH226 pKa = 5.74IRR228 pKa = 11.84IGDD231 pKa = 3.72LGVGQILHH239 pKa = 5.97VLVQGRR245 pKa = 11.84HH246 pKa = 4.93IVPEE250 pKa = 4.16DD251 pKa = 3.39VQLEE255 pKa = 4.32DD256 pKa = 4.71GVMQRR261 pKa = 11.84VKK263 pKa = 10.61IEE265 pKa = 3.77VGGDD269 pKa = 3.26DD270 pKa = 3.42VRR272 pKa = 11.84IDD274 pKa = 3.42VVRR277 pKa = 11.84RR278 pKa = 11.84MLDD281 pKa = 2.76RR282 pKa = 11.84GKK284 pKa = 9.93IEE286 pKa = 4.69HH287 pKa = 6.47IVIIRR292 pKa = 11.84HH293 pKa = 5.52NDD295 pKa = 2.59HH296 pKa = 7.0AARR299 pKa = 11.84VLTGGALDD307 pKa = 4.4PGAAFAQAVFLRR319 pKa = 11.84FMQGHH324 pKa = 5.92SPLVQVVFYY333 pKa = 10.36IAIGGLIGKK342 pKa = 7.93RR343 pKa = 11.84TGGPRR348 pKa = 11.84LKK350 pKa = 10.56DD351 pKa = 3.36VSLSEE356 pKa = 3.95QLLGVLVCLRR366 pKa = 11.84LVLTGKK372 pKa = 9.36VQVNIRR378 pKa = 11.84RR379 pKa = 11.84LVPVEE384 pKa = 3.62AQKK387 pKa = 10.96GFKK390 pKa = 10.13RR391 pKa = 11.84NVVTIPQIRR400 pKa = 11.84CATVFTVFRR409 pKa = 11.84RR410 pKa = 11.84QVKK413 pKa = 9.95ARR415 pKa = 11.84TDD417 pKa = 3.37RR418 pKa = 11.84AVGKK422 pKa = 9.36EE423 pKa = 3.94DD424 pKa = 3.5GVIAVWTAVVRR435 pKa = 11.84RR436 pKa = 11.84QRR438 pKa = 11.84IYY440 pKa = 11.11LRR442 pKa = 11.84DD443 pKa = 3.56ARR445 pKa = 11.84HH446 pKa = 6.57DD447 pKa = 3.79GDD449 pKa = 3.72KK450 pKa = 11.29GGTNRR455 pKa = 11.84TTRR458 pKa = 11.84AYY460 pKa = 10.54
MM1 pKa = 7.32QAVCCLFVLVGGILPVAQFVHH22 pKa = 6.46PQLDD26 pKa = 3.78HH27 pKa = 7.25LSALHH32 pKa = 6.45HH33 pKa = 5.95RR34 pKa = 11.84RR35 pKa = 11.84CVKK38 pKa = 10.11ILAAHH43 pKa = 6.52GKK45 pKa = 7.64VCRR48 pKa = 11.84IKK50 pKa = 10.44LQRR53 pKa = 11.84VVYY56 pKa = 8.92LVPAQANGSDD66 pKa = 4.42DD67 pKa = 2.99IRR69 pKa = 11.84CRR71 pKa = 11.84VRR73 pKa = 11.84LRR75 pKa = 11.84EE76 pKa = 4.33HH77 pKa = 6.68ILDD80 pKa = 4.48FKK82 pKa = 11.41AGVDD86 pKa = 3.55VPLRR90 pKa = 11.84HH91 pKa = 6.58IMLLHH96 pKa = 6.79LSDD99 pKa = 4.06VLLGQQLVCFSLSHH113 pKa = 6.31NLHH116 pKa = 5.94NFKK119 pKa = 10.22GHH121 pKa = 6.35RR122 pKa = 11.84RR123 pKa = 11.84VQPVVNQIQHH133 pKa = 6.58DD134 pKa = 4.33AVTGTDD140 pKa = 3.18NLADD144 pKa = 3.59VAGAAFDD151 pKa = 3.7EE152 pKa = 4.71VLRR155 pKa = 11.84IAQPYY160 pKa = 8.51VGAVGQTGNLQQIRR174 pKa = 11.84KK175 pKa = 8.73ILRR178 pKa = 11.84LTIQQHH184 pKa = 5.93LPDD187 pKa = 4.1KK188 pKa = 10.69RR189 pKa = 11.84RR190 pKa = 11.84AHH192 pKa = 6.27LRR194 pKa = 11.84DD195 pKa = 3.21GKK197 pKa = 10.96GAGFAVDD204 pKa = 4.88LLRR207 pKa = 11.84CHH209 pKa = 5.83TQHH212 pKa = 6.86LRR214 pKa = 11.84RR215 pKa = 11.84GEE217 pKa = 4.14QAVHH221 pKa = 6.07LLVRR225 pKa = 11.84HH226 pKa = 5.74IRR228 pKa = 11.84IGDD231 pKa = 3.72LGVGQILHH239 pKa = 5.97VLVQGRR245 pKa = 11.84HH246 pKa = 4.93IVPEE250 pKa = 4.16DD251 pKa = 3.39VQLEE255 pKa = 4.32DD256 pKa = 4.71GVMQRR261 pKa = 11.84VKK263 pKa = 10.61IEE265 pKa = 3.77VGGDD269 pKa = 3.26DD270 pKa = 3.42VRR272 pKa = 11.84IDD274 pKa = 3.42VVRR277 pKa = 11.84RR278 pKa = 11.84MLDD281 pKa = 2.76RR282 pKa = 11.84GKK284 pKa = 9.93IEE286 pKa = 4.69HH287 pKa = 6.47IVIIRR292 pKa = 11.84HH293 pKa = 5.52NDD295 pKa = 2.59HH296 pKa = 7.0AARR299 pKa = 11.84VLTGGALDD307 pKa = 4.4PGAAFAQAVFLRR319 pKa = 11.84FMQGHH324 pKa = 5.92SPLVQVVFYY333 pKa = 10.36IAIGGLIGKK342 pKa = 7.93RR343 pKa = 11.84TGGPRR348 pKa = 11.84LKK350 pKa = 10.56DD351 pKa = 3.36VSLSEE356 pKa = 3.95QLLGVLVCLRR366 pKa = 11.84LVLTGKK372 pKa = 9.36VQVNIRR378 pKa = 11.84RR379 pKa = 11.84LVPVEE384 pKa = 3.62AQKK387 pKa = 10.96GFKK390 pKa = 10.13RR391 pKa = 11.84NVVTIPQIRR400 pKa = 11.84CATVFTVFRR409 pKa = 11.84RR410 pKa = 11.84QVKK413 pKa = 9.95ARR415 pKa = 11.84TDD417 pKa = 3.37RR418 pKa = 11.84AVGKK422 pKa = 9.36EE423 pKa = 3.94DD424 pKa = 3.5GVIAVWTAVVRR435 pKa = 11.84RR436 pKa = 11.84QRR438 pKa = 11.84IYY440 pKa = 11.11LRR442 pKa = 11.84DD443 pKa = 3.56ARR445 pKa = 11.84HH446 pKa = 6.57DD447 pKa = 3.79GDD449 pKa = 3.72KK450 pKa = 11.29GGTNRR455 pKa = 11.84TTRR458 pKa = 11.84AYY460 pKa = 10.54
Molecular weight: 51.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
554686 |
30 |
1267 |
281.6 |
31.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.078 ± 0.055 | 1.829 ± 0.027 |
5.355 ± 0.046 | 6.936 ± 0.061 |
4.086 ± 0.04 | 7.195 ± 0.061 |
1.895 ± 0.026 | 6.304 ± 0.052 |
5.513 ± 0.049 | 9.981 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.991 ± 0.031 | 3.723 ± 0.038 |
3.791 ± 0.033 | 4.822 ± 0.054 |
4.942 ± 0.051 | 5.997 ± 0.05 |
5.141 ± 0.038 | 7.012 ± 0.051 |
0.895 ± 0.02 | 3.51 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
