
Beihai weivirus-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
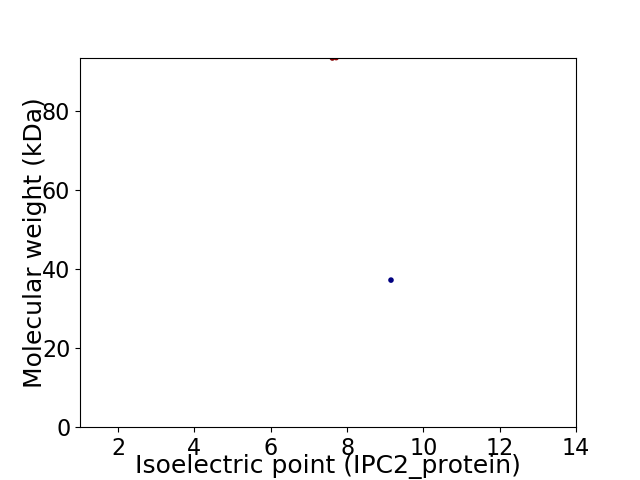
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL33|A0A1L3KL33_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 12 OX=1922741 PE=4 SV=1
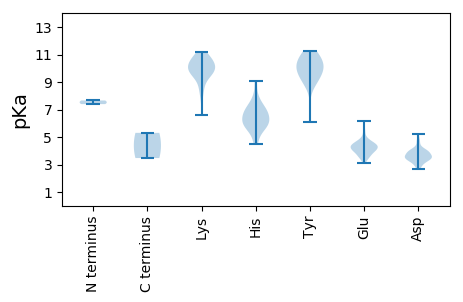
MM1 pKa = 7.39AVVSEE6 pKa = 4.48RR7 pKa = 11.84PSVSSVPLCPTCHH20 pKa = 6.17RR21 pKa = 11.84QHH23 pKa = 7.02LDD25 pKa = 3.16KK26 pKa = 11.15LSEE29 pKa = 3.69EE30 pKa = 4.43AAWDD34 pKa = 3.74RR35 pKa = 11.84ASKK38 pKa = 10.85DD39 pKa = 3.46EE40 pKa = 4.16SHH42 pKa = 5.87VVCVKK47 pKa = 8.82FHH49 pKa = 4.87VRR51 pKa = 11.84KK52 pKa = 9.64CAFCTDD58 pKa = 2.66VDD60 pKa = 4.03INCRR64 pKa = 11.84RR65 pKa = 11.84SFFSRR70 pKa = 11.84FTPRR74 pKa = 11.84TAHH77 pKa = 6.41SGDD80 pKa = 3.65SVLCEE85 pKa = 3.54VHH87 pKa = 5.62MTHH90 pKa = 6.47QEE92 pKa = 3.53LRR94 pKa = 11.84IARR97 pKa = 11.84DD98 pKa = 3.24AVGASLYY105 pKa = 10.77SEE107 pKa = 4.35APSRR111 pKa = 11.84VGNSNLHH118 pKa = 5.5LSGMRR123 pKa = 11.84LDD125 pKa = 4.78RR126 pKa = 11.84KK127 pKa = 10.66YY128 pKa = 11.25EE129 pKa = 3.89DD130 pKa = 4.21TEE132 pKa = 4.5MVWARR137 pKa = 11.84DD138 pKa = 3.6VVFPAVAIAMHH149 pKa = 7.25RR150 pKa = 11.84SRR152 pKa = 11.84EE153 pKa = 4.17TPVFSLGASIFPRR166 pKa = 11.84LRR168 pKa = 11.84KK169 pKa = 9.41LRR171 pKa = 11.84LEE173 pKa = 4.29SEE175 pKa = 4.44TGSKK179 pKa = 9.28RR180 pKa = 11.84WFWIRR185 pKa = 11.84ASQHH189 pKa = 4.48MTLEE193 pKa = 4.3CLRR196 pKa = 11.84FCPRR200 pKa = 11.84RR201 pKa = 11.84VEE203 pKa = 4.06QPTEE207 pKa = 3.66EE208 pKa = 4.61VYY210 pKa = 11.19APVATITPDD219 pKa = 3.07PGYY222 pKa = 9.12TFTRR226 pKa = 11.84FDD228 pKa = 3.64GVDD231 pKa = 3.25RR232 pKa = 11.84AATRR236 pKa = 11.84QRR238 pKa = 11.84EE239 pKa = 4.46GWDD242 pKa = 3.23LMASKK247 pKa = 10.93VFGVDD252 pKa = 4.08LMTDD256 pKa = 3.14AAGNPTLGDD265 pKa = 3.55NILAVRR271 pKa = 11.84HH272 pKa = 5.9LPFFARR278 pKa = 11.84KK279 pKa = 9.28IFFDD283 pKa = 3.48TLPRR287 pKa = 11.84NIAAAIAGRR296 pKa = 11.84IEE298 pKa = 4.5NVPPMQMTDD307 pKa = 3.5DD308 pKa = 3.9EE309 pKa = 4.42QAEE312 pKa = 4.2LRR314 pKa = 11.84GVTEE318 pKa = 4.42ALSICLRR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 2.84AKK329 pKa = 9.86MIRR332 pKa = 11.84EE333 pKa = 4.03IVTVLTLGDD342 pKa = 3.6WKK344 pKa = 10.76SKK346 pKa = 9.39KK347 pKa = 6.62WTITRR352 pKa = 11.84SKK354 pKa = 7.75TTLEE358 pKa = 4.03QLRR361 pKa = 11.84QRR363 pKa = 11.84YY364 pKa = 8.55NPTYY368 pKa = 9.93QFKK371 pKa = 10.6GQIKK375 pKa = 10.4LEE377 pKa = 4.01PSKK380 pKa = 10.52PGKK383 pKa = 9.38PPRR386 pKa = 11.84LIIADD391 pKa = 3.59GDD393 pKa = 4.01YY394 pKa = 10.12GQVMAWTIIATLEE407 pKa = 3.43RR408 pKa = 11.84WIFKK412 pKa = 10.09RR413 pKa = 11.84YY414 pKa = 6.07NHH416 pKa = 6.68RR417 pKa = 11.84SIKK420 pKa = 10.18GVCKK424 pKa = 10.03SDD426 pKa = 2.75AMKK429 pKa = 10.78RR430 pKa = 11.84LIKK433 pKa = 9.87NTAQFRR439 pKa = 11.84PSTGRR444 pKa = 11.84STTRR448 pKa = 11.84EE449 pKa = 3.56PVMVLEE455 pKa = 4.82NDD457 pKa = 3.32GSAWDD462 pKa = 3.61ACMSVALRR470 pKa = 11.84ALTEE474 pKa = 3.97NVLMQEE480 pKa = 4.06VHH482 pKa = 7.64DD483 pKa = 3.83ITKK486 pKa = 10.53DD487 pKa = 3.47LFIMEE492 pKa = 4.6SHH494 pKa = 6.3WQPEE498 pKa = 3.92RR499 pKa = 11.84LRR501 pKa = 11.84ANKK504 pKa = 9.66VKK506 pKa = 9.68TLSLSVAAKK515 pKa = 8.41FNCYY519 pKa = 9.85DD520 pKa = 3.26EE521 pKa = 5.62HH522 pKa = 9.08VPDD525 pKa = 4.76DD526 pKa = 3.86VRR528 pKa = 11.84NALAAGKK535 pKa = 8.14TYY537 pKa = 10.84KK538 pKa = 10.44EE539 pKa = 4.01VIAAIRR545 pKa = 11.84RR546 pKa = 11.84SGCRR550 pKa = 11.84GTSVLNFLANMILWCWVLGGKK571 pKa = 9.8DD572 pKa = 3.71GSRR575 pKa = 11.84LMEE578 pKa = 4.46SNGQKK583 pKa = 9.18FTDD586 pKa = 3.36VFGVVRR592 pKa = 11.84FVRR595 pKa = 11.84MMFEE599 pKa = 4.73GDD601 pKa = 4.07DD602 pKa = 3.85SLLTLTGARR611 pKa = 11.84FTPEE615 pKa = 3.27QVAQLSARR623 pKa = 11.84WTKK626 pKa = 10.48LGHH629 pKa = 5.76RR630 pKa = 11.84PKK632 pKa = 10.79LFVRR636 pKa = 11.84APGDD640 pKa = 3.3QAEE643 pKa = 4.24FTGYY647 pKa = 10.67KK648 pKa = 10.11FLVDD652 pKa = 3.37EE653 pKa = 5.38HH654 pKa = 8.01GLVPDD659 pKa = 4.44SEE661 pKa = 5.07VPDD664 pKa = 4.03LPRR667 pKa = 11.84LLGNVSYY674 pKa = 10.29CHH676 pKa = 6.38NRR678 pKa = 11.84GAVDD682 pKa = 3.38AAVKK686 pKa = 10.4GDD688 pKa = 3.44EE689 pKa = 4.36AALHH693 pKa = 5.98RR694 pKa = 11.84AVDD697 pKa = 3.8PGLLSRR703 pKa = 11.84AYY705 pKa = 9.27TIARR709 pKa = 11.84KK710 pKa = 9.85APTVARR716 pKa = 11.84WLYY719 pKa = 9.9NQVKK723 pKa = 8.57STDD726 pKa = 3.56LTFSNDD732 pKa = 2.86DD733 pKa = 3.51RR734 pKa = 11.84VRR736 pKa = 11.84LDD738 pKa = 4.42DD739 pKa = 5.23DD740 pKa = 4.76LGDD743 pKa = 4.0VLPEE747 pKa = 4.04LWKK750 pKa = 9.79GTDD753 pKa = 3.33GTEE756 pKa = 4.24KK757 pKa = 10.15MLEE760 pKa = 4.07QAISWQTFVEE770 pKa = 4.42RR771 pKa = 11.84VEE773 pKa = 4.44SEE775 pKa = 3.66ISAGEE780 pKa = 3.6AAGFDD785 pKa = 3.82EE786 pKa = 6.17AGFAYY791 pKa = 10.18AHH793 pKa = 6.67GWCTSTEE800 pKa = 3.68QWNDD804 pKa = 2.94FLIALGAITRR814 pKa = 11.84GTDD817 pKa = 3.22SVVVGRR823 pKa = 11.84VLPSCFF829 pKa = 3.47
MM1 pKa = 7.39AVVSEE6 pKa = 4.48RR7 pKa = 11.84PSVSSVPLCPTCHH20 pKa = 6.17RR21 pKa = 11.84QHH23 pKa = 7.02LDD25 pKa = 3.16KK26 pKa = 11.15LSEE29 pKa = 3.69EE30 pKa = 4.43AAWDD34 pKa = 3.74RR35 pKa = 11.84ASKK38 pKa = 10.85DD39 pKa = 3.46EE40 pKa = 4.16SHH42 pKa = 5.87VVCVKK47 pKa = 8.82FHH49 pKa = 4.87VRR51 pKa = 11.84KK52 pKa = 9.64CAFCTDD58 pKa = 2.66VDD60 pKa = 4.03INCRR64 pKa = 11.84RR65 pKa = 11.84SFFSRR70 pKa = 11.84FTPRR74 pKa = 11.84TAHH77 pKa = 6.41SGDD80 pKa = 3.65SVLCEE85 pKa = 3.54VHH87 pKa = 5.62MTHH90 pKa = 6.47QEE92 pKa = 3.53LRR94 pKa = 11.84IARR97 pKa = 11.84DD98 pKa = 3.24AVGASLYY105 pKa = 10.77SEE107 pKa = 4.35APSRR111 pKa = 11.84VGNSNLHH118 pKa = 5.5LSGMRR123 pKa = 11.84LDD125 pKa = 4.78RR126 pKa = 11.84KK127 pKa = 10.66YY128 pKa = 11.25EE129 pKa = 3.89DD130 pKa = 4.21TEE132 pKa = 4.5MVWARR137 pKa = 11.84DD138 pKa = 3.6VVFPAVAIAMHH149 pKa = 7.25RR150 pKa = 11.84SRR152 pKa = 11.84EE153 pKa = 4.17TPVFSLGASIFPRR166 pKa = 11.84LRR168 pKa = 11.84KK169 pKa = 9.41LRR171 pKa = 11.84LEE173 pKa = 4.29SEE175 pKa = 4.44TGSKK179 pKa = 9.28RR180 pKa = 11.84WFWIRR185 pKa = 11.84ASQHH189 pKa = 4.48MTLEE193 pKa = 4.3CLRR196 pKa = 11.84FCPRR200 pKa = 11.84RR201 pKa = 11.84VEE203 pKa = 4.06QPTEE207 pKa = 3.66EE208 pKa = 4.61VYY210 pKa = 11.19APVATITPDD219 pKa = 3.07PGYY222 pKa = 9.12TFTRR226 pKa = 11.84FDD228 pKa = 3.64GVDD231 pKa = 3.25RR232 pKa = 11.84AATRR236 pKa = 11.84QRR238 pKa = 11.84EE239 pKa = 4.46GWDD242 pKa = 3.23LMASKK247 pKa = 10.93VFGVDD252 pKa = 4.08LMTDD256 pKa = 3.14AAGNPTLGDD265 pKa = 3.55NILAVRR271 pKa = 11.84HH272 pKa = 5.9LPFFARR278 pKa = 11.84KK279 pKa = 9.28IFFDD283 pKa = 3.48TLPRR287 pKa = 11.84NIAAAIAGRR296 pKa = 11.84IEE298 pKa = 4.5NVPPMQMTDD307 pKa = 3.5DD308 pKa = 3.9EE309 pKa = 4.42QAEE312 pKa = 4.2LRR314 pKa = 11.84GVTEE318 pKa = 4.42ALSICLRR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 2.84AKK329 pKa = 9.86MIRR332 pKa = 11.84EE333 pKa = 4.03IVTVLTLGDD342 pKa = 3.6WKK344 pKa = 10.76SKK346 pKa = 9.39KK347 pKa = 6.62WTITRR352 pKa = 11.84SKK354 pKa = 7.75TTLEE358 pKa = 4.03QLRR361 pKa = 11.84QRR363 pKa = 11.84YY364 pKa = 8.55NPTYY368 pKa = 9.93QFKK371 pKa = 10.6GQIKK375 pKa = 10.4LEE377 pKa = 4.01PSKK380 pKa = 10.52PGKK383 pKa = 9.38PPRR386 pKa = 11.84LIIADD391 pKa = 3.59GDD393 pKa = 4.01YY394 pKa = 10.12GQVMAWTIIATLEE407 pKa = 3.43RR408 pKa = 11.84WIFKK412 pKa = 10.09RR413 pKa = 11.84YY414 pKa = 6.07NHH416 pKa = 6.68RR417 pKa = 11.84SIKK420 pKa = 10.18GVCKK424 pKa = 10.03SDD426 pKa = 2.75AMKK429 pKa = 10.78RR430 pKa = 11.84LIKK433 pKa = 9.87NTAQFRR439 pKa = 11.84PSTGRR444 pKa = 11.84STTRR448 pKa = 11.84EE449 pKa = 3.56PVMVLEE455 pKa = 4.82NDD457 pKa = 3.32GSAWDD462 pKa = 3.61ACMSVALRR470 pKa = 11.84ALTEE474 pKa = 3.97NVLMQEE480 pKa = 4.06VHH482 pKa = 7.64DD483 pKa = 3.83ITKK486 pKa = 10.53DD487 pKa = 3.47LFIMEE492 pKa = 4.6SHH494 pKa = 6.3WQPEE498 pKa = 3.92RR499 pKa = 11.84LRR501 pKa = 11.84ANKK504 pKa = 9.66VKK506 pKa = 9.68TLSLSVAAKK515 pKa = 8.41FNCYY519 pKa = 9.85DD520 pKa = 3.26EE521 pKa = 5.62HH522 pKa = 9.08VPDD525 pKa = 4.76DD526 pKa = 3.86VRR528 pKa = 11.84NALAAGKK535 pKa = 8.14TYY537 pKa = 10.84KK538 pKa = 10.44EE539 pKa = 4.01VIAAIRR545 pKa = 11.84RR546 pKa = 11.84SGCRR550 pKa = 11.84GTSVLNFLANMILWCWVLGGKK571 pKa = 9.8DD572 pKa = 3.71GSRR575 pKa = 11.84LMEE578 pKa = 4.46SNGQKK583 pKa = 9.18FTDD586 pKa = 3.36VFGVVRR592 pKa = 11.84FVRR595 pKa = 11.84MMFEE599 pKa = 4.73GDD601 pKa = 4.07DD602 pKa = 3.85SLLTLTGARR611 pKa = 11.84FTPEE615 pKa = 3.27QVAQLSARR623 pKa = 11.84WTKK626 pKa = 10.48LGHH629 pKa = 5.76RR630 pKa = 11.84PKK632 pKa = 10.79LFVRR636 pKa = 11.84APGDD640 pKa = 3.3QAEE643 pKa = 4.24FTGYY647 pKa = 10.67KK648 pKa = 10.11FLVDD652 pKa = 3.37EE653 pKa = 5.38HH654 pKa = 8.01GLVPDD659 pKa = 4.44SEE661 pKa = 5.07VPDD664 pKa = 4.03LPRR667 pKa = 11.84LLGNVSYY674 pKa = 10.29CHH676 pKa = 6.38NRR678 pKa = 11.84GAVDD682 pKa = 3.38AAVKK686 pKa = 10.4GDD688 pKa = 3.44EE689 pKa = 4.36AALHH693 pKa = 5.98RR694 pKa = 11.84AVDD697 pKa = 3.8PGLLSRR703 pKa = 11.84AYY705 pKa = 9.27TIARR709 pKa = 11.84KK710 pKa = 9.85APTVARR716 pKa = 11.84WLYY719 pKa = 9.9NQVKK723 pKa = 8.57STDD726 pKa = 3.56LTFSNDD732 pKa = 2.86DD733 pKa = 3.51RR734 pKa = 11.84VRR736 pKa = 11.84LDD738 pKa = 4.42DD739 pKa = 5.23DD740 pKa = 4.76LGDD743 pKa = 4.0VLPEE747 pKa = 4.04LWKK750 pKa = 9.79GTDD753 pKa = 3.33GTEE756 pKa = 4.24KK757 pKa = 10.15MLEE760 pKa = 4.07QAISWQTFVEE770 pKa = 4.42RR771 pKa = 11.84VEE773 pKa = 4.44SEE775 pKa = 3.66ISAGEE780 pKa = 3.6AAGFDD785 pKa = 3.82EE786 pKa = 6.17AGFAYY791 pKa = 10.18AHH793 pKa = 6.67GWCTSTEE800 pKa = 3.68QWNDD804 pKa = 2.94FLIALGAITRR814 pKa = 11.84GTDD817 pKa = 3.22SVVVGRR823 pKa = 11.84VLPSCFF829 pKa = 3.47
Molecular weight: 93.37 kDa
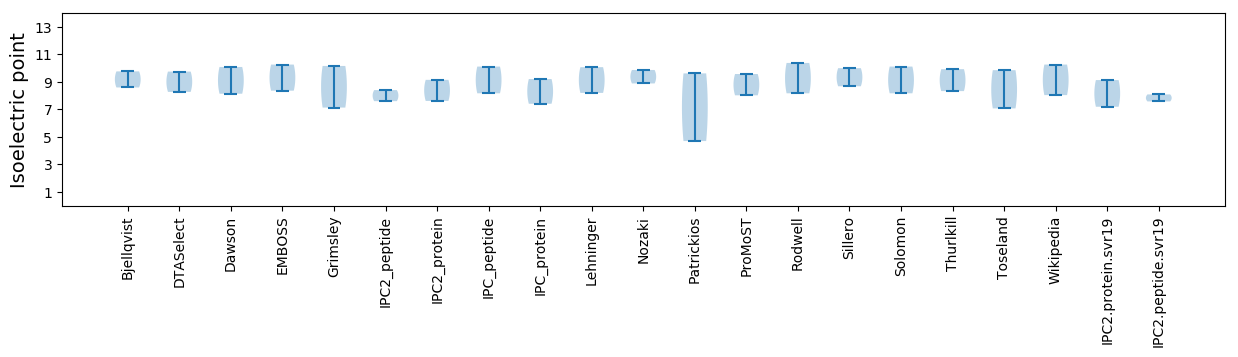
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL33|A0A1L3KL33_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 12 OX=1922741 PE=4 SV=1
MM1 pKa = 7.66PASKK5 pKa = 9.87SQRR8 pKa = 11.84VRR10 pKa = 11.84AVMKK14 pKa = 10.44KK15 pKa = 10.24ALKK18 pKa = 10.07KK19 pKa = 10.27KK20 pKa = 10.24QGGANLYY27 pKa = 10.18GIKK30 pKa = 10.01QGAAMVNTRR39 pKa = 11.84PFPRR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84NGRR50 pKa = 11.84GSSDD54 pKa = 2.97ARR56 pKa = 11.84LATMIKK62 pKa = 9.74YY63 pKa = 9.81GLNALHH69 pKa = 7.11PCHH72 pKa = 7.02LALPRR77 pKa = 11.84AVGGYY82 pKa = 9.13SVVRR86 pKa = 11.84TTDD89 pKa = 3.48IINSNAATMFFGTFKK104 pKa = 11.21GPGNQFTEE112 pKa = 4.77TTWLSTIAIRR122 pKa = 11.84GVAPDD127 pKa = 3.49TAINGTGNAFLYY139 pKa = 10.73NSSALQAPSLSGTRR153 pKa = 11.84MVPAAMTVQIMNGQSLQNADD173 pKa = 3.71GIAYY177 pKa = 9.27IGKK180 pKa = 9.69SKK182 pKa = 9.6TVLDD186 pKa = 4.01LMGDD190 pKa = 3.35TRR192 pKa = 11.84TWRR195 pKa = 11.84TVFEE199 pKa = 4.7DD200 pKa = 3.83LVSYY204 pKa = 9.24SAPRR208 pKa = 11.84LCSGGKK214 pKa = 8.87LALRR218 pKa = 11.84GVQVDD223 pKa = 3.9AVPNNLSVLSDD234 pKa = 3.65FVPRR238 pKa = 11.84RR239 pKa = 11.84IPTEE243 pKa = 4.05GARR246 pKa = 11.84TWDD249 pKa = 3.66EE250 pKa = 3.91DD251 pKa = 3.44QAALDD256 pKa = 4.09FEE258 pKa = 4.81GFAPIFVYY266 pKa = 10.8NPDD269 pKa = 4.03NIDD272 pKa = 3.57LKK274 pKa = 11.22FLVTIEE280 pKa = 3.09WRR282 pKa = 11.84MRR284 pKa = 11.84FDD286 pKa = 4.19PLNPAYY292 pKa = 10.0AGHH295 pKa = 5.73VTHH298 pKa = 7.08TPAAEE303 pKa = 4.33STWSKK308 pKa = 10.97VLAHH312 pKa = 6.74AEE314 pKa = 4.23SLGHH318 pKa = 5.95GVMDD322 pKa = 3.97IADD325 pKa = 3.58VVADD329 pKa = 3.92VGAAGLRR336 pKa = 11.84ARR338 pKa = 11.84DD339 pKa = 3.92AIAPLLAA346 pKa = 5.3
MM1 pKa = 7.66PASKK5 pKa = 9.87SQRR8 pKa = 11.84VRR10 pKa = 11.84AVMKK14 pKa = 10.44KK15 pKa = 10.24ALKK18 pKa = 10.07KK19 pKa = 10.27KK20 pKa = 10.24QGGANLYY27 pKa = 10.18GIKK30 pKa = 10.01QGAAMVNTRR39 pKa = 11.84PFPRR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84NGRR50 pKa = 11.84GSSDD54 pKa = 2.97ARR56 pKa = 11.84LATMIKK62 pKa = 9.74YY63 pKa = 9.81GLNALHH69 pKa = 7.11PCHH72 pKa = 7.02LALPRR77 pKa = 11.84AVGGYY82 pKa = 9.13SVVRR86 pKa = 11.84TTDD89 pKa = 3.48IINSNAATMFFGTFKK104 pKa = 11.21GPGNQFTEE112 pKa = 4.77TTWLSTIAIRR122 pKa = 11.84GVAPDD127 pKa = 3.49TAINGTGNAFLYY139 pKa = 10.73NSSALQAPSLSGTRR153 pKa = 11.84MVPAAMTVQIMNGQSLQNADD173 pKa = 3.71GIAYY177 pKa = 9.27IGKK180 pKa = 9.69SKK182 pKa = 9.6TVLDD186 pKa = 4.01LMGDD190 pKa = 3.35TRR192 pKa = 11.84TWRR195 pKa = 11.84TVFEE199 pKa = 4.7DD200 pKa = 3.83LVSYY204 pKa = 9.24SAPRR208 pKa = 11.84LCSGGKK214 pKa = 8.87LALRR218 pKa = 11.84GVQVDD223 pKa = 3.9AVPNNLSVLSDD234 pKa = 3.65FVPRR238 pKa = 11.84RR239 pKa = 11.84IPTEE243 pKa = 4.05GARR246 pKa = 11.84TWDD249 pKa = 3.66EE250 pKa = 3.91DD251 pKa = 3.44QAALDD256 pKa = 4.09FEE258 pKa = 4.81GFAPIFVYY266 pKa = 10.8NPDD269 pKa = 4.03NIDD272 pKa = 3.57LKK274 pKa = 11.22FLVTIEE280 pKa = 3.09WRR282 pKa = 11.84MRR284 pKa = 11.84FDD286 pKa = 4.19PLNPAYY292 pKa = 10.0AGHH295 pKa = 5.73VTHH298 pKa = 7.08TPAAEE303 pKa = 4.33STWSKK308 pKa = 10.97VLAHH312 pKa = 6.74AEE314 pKa = 4.23SLGHH318 pKa = 5.95GVMDD322 pKa = 3.97IADD325 pKa = 3.58VVADD329 pKa = 3.92VGAAGLRR336 pKa = 11.84ARR338 pKa = 11.84DD339 pKa = 3.92AIAPLLAA346 pKa = 5.3
Molecular weight: 37.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1175 |
346 |
829 |
587.5 |
65.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.128 ± 1.59 | 1.702 ± 0.621 |
6.213 ± 0.399 | 4.851 ± 1.403 |
4.255 ± 0.275 | 6.809 ± 1.029 |
2.213 ± 0.264 | 4.17 ± 0.251 |
4.511 ± 0.257 | 8.426 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.809 ± 0.205 | 3.404 ± 0.834 |
4.766 ± 0.401 | 2.809 ± 0.045 |
8.17 ± 0.682 | 6.298 ± 0.126 |
6.723 ± 0.118 | 7.745 ± 0.287 |
2.043 ± 0.33 | 1.957 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
