
Zinderia insecticola (strain CARI)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Candidatus Zinderia; Candidatus Zinderia insecticola
Average proteome isoelectric point is 9.02
Get precalculated fractions of proteins
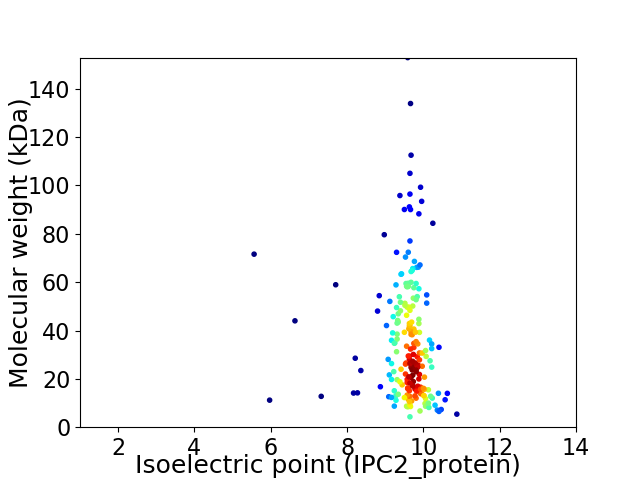
Virtual 2D-PAGE plot for 206 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
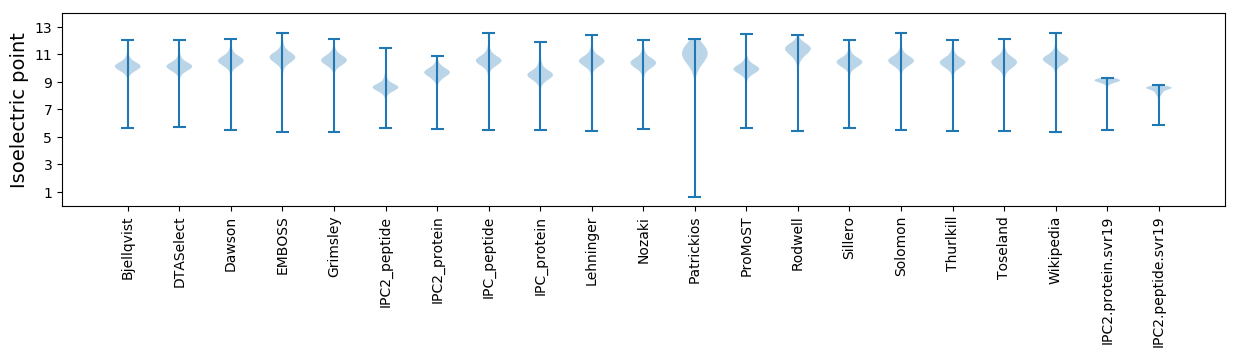
* You can choose from 21 different methods for calculating isoelectric point
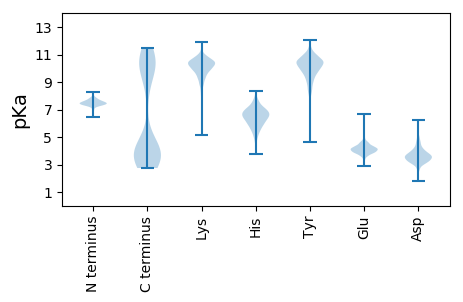
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0TIY9|E0TIY9_ZINIC Protein GrpE OS=Zinderia insecticola (strain CARI) OX=871271 GN=grpE PE=3 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCVSIMEE20 pKa = 4.42GNQSKK25 pKa = 10.82VIEE28 pKa = 4.24NSEE31 pKa = 4.17GSRR34 pKa = 11.84TTPSVVAYY42 pKa = 9.35QEE44 pKa = 3.85NGEE47 pKa = 4.28ILVGSPAKK55 pKa = 10.2RR56 pKa = 11.84QAITNPKK63 pKa = 7.29NTIYY67 pKa = 10.56AAKK70 pKa = 10.01RR71 pKa = 11.84LIGRR75 pKa = 11.84KK76 pKa = 8.68FNEE79 pKa = 4.23KK80 pKa = 9.68EE81 pKa = 3.97VQKK84 pKa = 10.87DD85 pKa = 3.18IDD87 pKa = 3.57IMPYY91 pKa = 10.41EE92 pKa = 4.61IIKK95 pKa = 10.61SNNGDD100 pKa = 2.57AWINIRR106 pKa = 11.84NKK108 pKa = 10.26KK109 pKa = 8.65IAPPQISAEE118 pKa = 4.18VLKK121 pKa = 10.99KK122 pKa = 9.74MKK124 pKa = 9.1KK125 pKa = 7.18TAEE128 pKa = 4.41DD129 pKa = 3.66YY130 pKa = 11.45LGFKK134 pKa = 10.41VNEE137 pKa = 4.05AVITVPAYY145 pKa = 10.64FNDD148 pKa = 3.82AQRR151 pKa = 11.84QATKK155 pKa = 10.58DD156 pKa = 3.07AGRR159 pKa = 11.84IAGLEE164 pKa = 3.98VKK166 pKa = 10.42RR167 pKa = 11.84IINEE171 pKa = 3.62PTAAALAFGLDD182 pKa = 3.21KK183 pKa = 11.23VLNNDD188 pKa = 3.38KK189 pKa = 10.79KK190 pKa = 10.67IAVYY194 pKa = 10.58DD195 pKa = 3.97LGGGTFDD202 pKa = 3.69ISIIEE207 pKa = 3.79ISNIEE212 pKa = 4.02GEE214 pKa = 4.33KK215 pKa = 10.16QFEE218 pKa = 4.28VLSTNGDD225 pKa = 3.61TFLGGEE231 pKa = 4.52DD232 pKa = 4.01FDD234 pKa = 5.48KK235 pKa = 11.37KK236 pKa = 11.08LIDD239 pKa = 3.59YY240 pKa = 8.47VINEE244 pKa = 4.15FKK246 pKa = 10.84KK247 pKa = 10.74NNGIDD252 pKa = 3.64LYY254 pKa = 10.58KK255 pKa = 10.94DD256 pKa = 3.54SIALQRR262 pKa = 11.84IKK264 pKa = 11.08SSVEE268 pKa = 3.37RR269 pKa = 11.84AKK271 pKa = 10.58IEE273 pKa = 3.98LSSLYY278 pKa = 8.58EE279 pKa = 3.94TEE281 pKa = 3.87INEE284 pKa = 4.72PYY286 pKa = 9.25ITVNNNIPLHH296 pKa = 6.41LNLKK300 pKa = 7.58ITRR303 pKa = 11.84SKK305 pKa = 11.0FEE307 pKa = 4.34SLVEE311 pKa = 3.85DD312 pKa = 5.66LINKK316 pKa = 7.35TIKK319 pKa = 9.77PCEE322 pKa = 3.97IAVKK326 pKa = 10.01DD327 pKa = 3.59AKK329 pKa = 10.89ININEE334 pKa = 3.97ISDD337 pKa = 4.06VILVGGMTRR346 pKa = 11.84MPKK349 pKa = 9.5IQEE352 pKa = 3.81KK353 pKa = 10.83VKK355 pKa = 10.6IFFNKK360 pKa = 9.8EE361 pKa = 3.34PRR363 pKa = 11.84KK364 pKa = 10.42DD365 pKa = 3.57INPDD369 pKa = 2.89EE370 pKa = 4.35AVAIGAAIQGAVISGDD386 pKa = 3.66KK387 pKa = 10.45KK388 pKa = 11.05DD389 pKa = 5.02LLLLDD394 pKa = 3.97VTPLSLGIEE403 pKa = 4.29TMGGIMTKK411 pKa = 9.83MIKK414 pKa = 10.32KK415 pKa = 8.22NTTIPTKK422 pKa = 10.43FSQVFSTAEE431 pKa = 3.91DD432 pKa = 3.65NQPMVTIRR440 pKa = 11.84IYY442 pKa = 10.02QGEE445 pKa = 4.08RR446 pKa = 11.84EE447 pKa = 4.02MANNNKK453 pKa = 9.84LLGEE457 pKa = 4.27FNLEE461 pKa = 4.54GIEE464 pKa = 4.16SAPRR468 pKa = 11.84GVPQIEE474 pKa = 4.17VTFDD478 pKa = 2.89IDD480 pKa = 3.83VNGILHH486 pKa = 5.51VTAKK490 pKa = 10.65DD491 pKa = 3.26KK492 pKa = 10.17NTNKK496 pKa = 9.92EE497 pKa = 3.71NKK499 pKa = 8.35ITIKK503 pKa = 10.79ANSGLNEE510 pKa = 3.9NEE512 pKa = 3.66IEE514 pKa = 4.89KK515 pKa = 9.79MIKK518 pKa = 9.97DD519 pKa = 3.51AEE521 pKa = 4.38INSEE525 pKa = 4.05EE526 pKa = 4.12DD527 pKa = 3.43KK528 pKa = 10.96KK529 pKa = 11.17IKK531 pKa = 10.45DD532 pKa = 3.88LIEE535 pKa = 4.24VKK537 pKa = 10.78NKK539 pKa = 10.35ADD541 pKa = 3.9TLIHH545 pKa = 6.02SANKK549 pKa = 9.9IIEE552 pKa = 4.37KK553 pKa = 10.03NSNLISEE560 pKa = 4.26NDD562 pKa = 3.11INNLKK567 pKa = 10.59KK568 pKa = 10.62EE569 pKa = 4.13IIEE572 pKa = 4.59LEE574 pKa = 4.19DD575 pKa = 3.95LVKK578 pKa = 11.17LNDD581 pKa = 4.15KK582 pKa = 10.7IKK584 pKa = 10.49IEE586 pKa = 4.01NKK588 pKa = 9.48INSLSVILQKK598 pKa = 10.56INSEE602 pKa = 3.76IYY604 pKa = 10.06KK605 pKa = 10.42NKK607 pKa = 10.43NINKK611 pKa = 9.34KK612 pKa = 10.75DD613 pKa = 3.47EE614 pKa = 4.87DD615 pKa = 3.55IKK617 pKa = 11.68NNNEE621 pKa = 3.9DD622 pKa = 4.47LEE624 pKa = 4.92KK625 pKa = 10.83NDD627 pKa = 3.44EE628 pKa = 4.24NKK630 pKa = 9.86VVDD633 pKa = 5.23KK634 pKa = 10.95DD635 pKa = 4.04FKK637 pKa = 11.02DD638 pKa = 3.59IKK640 pKa = 10.94
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCVSIMEE20 pKa = 4.42GNQSKK25 pKa = 10.82VIEE28 pKa = 4.24NSEE31 pKa = 4.17GSRR34 pKa = 11.84TTPSVVAYY42 pKa = 9.35QEE44 pKa = 3.85NGEE47 pKa = 4.28ILVGSPAKK55 pKa = 10.2RR56 pKa = 11.84QAITNPKK63 pKa = 7.29NTIYY67 pKa = 10.56AAKK70 pKa = 10.01RR71 pKa = 11.84LIGRR75 pKa = 11.84KK76 pKa = 8.68FNEE79 pKa = 4.23KK80 pKa = 9.68EE81 pKa = 3.97VQKK84 pKa = 10.87DD85 pKa = 3.18IDD87 pKa = 3.57IMPYY91 pKa = 10.41EE92 pKa = 4.61IIKK95 pKa = 10.61SNNGDD100 pKa = 2.57AWINIRR106 pKa = 11.84NKK108 pKa = 10.26KK109 pKa = 8.65IAPPQISAEE118 pKa = 4.18VLKK121 pKa = 10.99KK122 pKa = 9.74MKK124 pKa = 9.1KK125 pKa = 7.18TAEE128 pKa = 4.41DD129 pKa = 3.66YY130 pKa = 11.45LGFKK134 pKa = 10.41VNEE137 pKa = 4.05AVITVPAYY145 pKa = 10.64FNDD148 pKa = 3.82AQRR151 pKa = 11.84QATKK155 pKa = 10.58DD156 pKa = 3.07AGRR159 pKa = 11.84IAGLEE164 pKa = 3.98VKK166 pKa = 10.42RR167 pKa = 11.84IINEE171 pKa = 3.62PTAAALAFGLDD182 pKa = 3.21KK183 pKa = 11.23VLNNDD188 pKa = 3.38KK189 pKa = 10.79KK190 pKa = 10.67IAVYY194 pKa = 10.58DD195 pKa = 3.97LGGGTFDD202 pKa = 3.69ISIIEE207 pKa = 3.79ISNIEE212 pKa = 4.02GEE214 pKa = 4.33KK215 pKa = 10.16QFEE218 pKa = 4.28VLSTNGDD225 pKa = 3.61TFLGGEE231 pKa = 4.52DD232 pKa = 4.01FDD234 pKa = 5.48KK235 pKa = 11.37KK236 pKa = 11.08LIDD239 pKa = 3.59YY240 pKa = 8.47VINEE244 pKa = 4.15FKK246 pKa = 10.84KK247 pKa = 10.74NNGIDD252 pKa = 3.64LYY254 pKa = 10.58KK255 pKa = 10.94DD256 pKa = 3.54SIALQRR262 pKa = 11.84IKK264 pKa = 11.08SSVEE268 pKa = 3.37RR269 pKa = 11.84AKK271 pKa = 10.58IEE273 pKa = 3.98LSSLYY278 pKa = 8.58EE279 pKa = 3.94TEE281 pKa = 3.87INEE284 pKa = 4.72PYY286 pKa = 9.25ITVNNNIPLHH296 pKa = 6.41LNLKK300 pKa = 7.58ITRR303 pKa = 11.84SKK305 pKa = 11.0FEE307 pKa = 4.34SLVEE311 pKa = 3.85DD312 pKa = 5.66LINKK316 pKa = 7.35TIKK319 pKa = 9.77PCEE322 pKa = 3.97IAVKK326 pKa = 10.01DD327 pKa = 3.59AKK329 pKa = 10.89ININEE334 pKa = 3.97ISDD337 pKa = 4.06VILVGGMTRR346 pKa = 11.84MPKK349 pKa = 9.5IQEE352 pKa = 3.81KK353 pKa = 10.83VKK355 pKa = 10.6IFFNKK360 pKa = 9.8EE361 pKa = 3.34PRR363 pKa = 11.84KK364 pKa = 10.42DD365 pKa = 3.57INPDD369 pKa = 2.89EE370 pKa = 4.35AVAIGAAIQGAVISGDD386 pKa = 3.66KK387 pKa = 10.45KK388 pKa = 11.05DD389 pKa = 5.02LLLLDD394 pKa = 3.97VTPLSLGIEE403 pKa = 4.29TMGGIMTKK411 pKa = 9.83MIKK414 pKa = 10.32KK415 pKa = 8.22NTTIPTKK422 pKa = 10.43FSQVFSTAEE431 pKa = 3.91DD432 pKa = 3.65NQPMVTIRR440 pKa = 11.84IYY442 pKa = 10.02QGEE445 pKa = 4.08RR446 pKa = 11.84EE447 pKa = 4.02MANNNKK453 pKa = 9.84LLGEE457 pKa = 4.27FNLEE461 pKa = 4.54GIEE464 pKa = 4.16SAPRR468 pKa = 11.84GVPQIEE474 pKa = 4.17VTFDD478 pKa = 2.89IDD480 pKa = 3.83VNGILHH486 pKa = 5.51VTAKK490 pKa = 10.65DD491 pKa = 3.26KK492 pKa = 10.17NTNKK496 pKa = 9.92EE497 pKa = 3.71NKK499 pKa = 8.35ITIKK503 pKa = 10.79ANSGLNEE510 pKa = 3.9NEE512 pKa = 3.66IEE514 pKa = 4.89KK515 pKa = 9.79MIKK518 pKa = 9.97DD519 pKa = 3.51AEE521 pKa = 4.38INSEE525 pKa = 4.05EE526 pKa = 4.12DD527 pKa = 3.43KK528 pKa = 10.96KK529 pKa = 11.17IKK531 pKa = 10.45DD532 pKa = 3.88LIEE535 pKa = 4.24VKK537 pKa = 10.78NKK539 pKa = 10.35ADD541 pKa = 3.9TLIHH545 pKa = 6.02SANKK549 pKa = 9.9IIEE552 pKa = 4.37KK553 pKa = 10.03NSNLISEE560 pKa = 4.26NDD562 pKa = 3.11INNLKK567 pKa = 10.59KK568 pKa = 10.62EE569 pKa = 4.13IIEE572 pKa = 4.59LEE574 pKa = 4.19DD575 pKa = 3.95LVKK578 pKa = 11.17LNDD581 pKa = 4.15KK582 pKa = 10.7IKK584 pKa = 10.49IEE586 pKa = 4.01NKK588 pKa = 9.48INSLSVILQKK598 pKa = 10.56INSEE602 pKa = 3.76IYY604 pKa = 10.06KK605 pKa = 10.42NKK607 pKa = 10.43NINKK611 pKa = 9.34KK612 pKa = 10.75DD613 pKa = 3.47EE614 pKa = 4.87DD615 pKa = 3.55IKK617 pKa = 11.68NNNEE621 pKa = 3.9DD622 pKa = 4.47LEE624 pKa = 4.92KK625 pKa = 10.83NDD627 pKa = 3.44EE628 pKa = 4.24NKK630 pKa = 9.86VVDD633 pKa = 5.23KK634 pKa = 10.95DD635 pKa = 4.04FKK637 pKa = 11.02DD638 pKa = 3.59IKK640 pKa = 10.94
Molecular weight: 71.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0TJ45|E0TJ45_ZINIC 50S ribosomal protein L4 OS=Zinderia insecticola (strain CARI) OX=871271 GN=rplD PE=3 SV=1
MM1 pKa = 7.75IIYY4 pKa = 9.64KK5 pKa = 9.54PKK7 pKa = 7.95PTSPGRR13 pKa = 11.84RR14 pKa = 11.84NVIKK18 pKa = 10.48ILNKK22 pKa = 9.69NLYY25 pKa = 9.13KK26 pKa = 10.66GRR28 pKa = 11.84PLSILLKK35 pKa = 10.2KK36 pKa = 10.09KK37 pKa = 9.9KK38 pKa = 10.02KK39 pKa = 9.88KK40 pKa = 9.96SGRR43 pKa = 11.84NNMGRR48 pKa = 11.84ITTRR52 pKa = 11.84HH53 pKa = 6.2KK54 pKa = 10.47GGQHH58 pKa = 4.35KK59 pKa = 9.92RR60 pKa = 11.84YY61 pKa = 10.13YY62 pKa = 10.45RR63 pKa = 11.84IIDD66 pKa = 3.88FKK68 pKa = 11.13RR69 pKa = 11.84NEE71 pKa = 4.09YY72 pKa = 10.0GIKK75 pKa = 10.4GYY77 pKa = 8.94VKK79 pKa = 10.26RR80 pKa = 11.84IEE82 pKa = 3.97YY83 pKa = 10.47DD84 pKa = 3.15PNRR87 pKa = 11.84NCNIALIYY95 pKa = 9.37YY96 pKa = 10.35LNGKK100 pKa = 8.7KK101 pKa = 10.07KK102 pKa = 10.93YY103 pKa = 9.63ILHH106 pKa = 6.7PKK108 pKa = 8.87NLNINDD114 pKa = 4.36EE115 pKa = 4.52IISNYY120 pKa = 7.41KK121 pKa = 9.2TPIKK125 pKa = 10.23IGNNLLIKK133 pKa = 10.11NIPLGTLIHH142 pKa = 6.44CLEE145 pKa = 4.56LLPFKK150 pKa = 10.45GAQIARR156 pKa = 11.84SAGSSALIISRR167 pKa = 11.84EE168 pKa = 3.78KK169 pKa = 10.65KK170 pKa = 8.08YY171 pKa = 8.72TQVRR175 pKa = 11.84LKK177 pKa = 10.6SNEE180 pKa = 3.02IRR182 pKa = 11.84RR183 pKa = 11.84INNNCFATIGEE194 pKa = 4.4IGNEE198 pKa = 3.59KK199 pKa = 10.58HH200 pKa = 6.83NLIKK204 pKa = 10.6LGKK207 pKa = 10.11AGIKK211 pKa = 9.83RR212 pKa = 11.84NLGIRR217 pKa = 11.84PTVRR221 pKa = 11.84GVAMNPIDD229 pKa = 4.06HH230 pKa = 6.89PHH232 pKa = 6.54GGGEE236 pKa = 4.02GRR238 pKa = 11.84TSGGRR243 pKa = 11.84HH244 pKa = 5.87PVTPWGKK251 pKa = 8.02KK252 pKa = 6.7TKK254 pKa = 10.06GKK256 pKa = 8.43KK257 pKa = 7.59TRR259 pKa = 11.84NKK261 pKa = 10.49KK262 pKa = 10.26KK263 pKa = 9.82ISNNMIINNKK273 pKa = 7.88NDD275 pKa = 3.56KK276 pKa = 10.67IFF278 pKa = 3.77
MM1 pKa = 7.75IIYY4 pKa = 9.64KK5 pKa = 9.54PKK7 pKa = 7.95PTSPGRR13 pKa = 11.84RR14 pKa = 11.84NVIKK18 pKa = 10.48ILNKK22 pKa = 9.69NLYY25 pKa = 9.13KK26 pKa = 10.66GRR28 pKa = 11.84PLSILLKK35 pKa = 10.2KK36 pKa = 10.09KK37 pKa = 9.9KK38 pKa = 10.02KK39 pKa = 9.88KK40 pKa = 9.96SGRR43 pKa = 11.84NNMGRR48 pKa = 11.84ITTRR52 pKa = 11.84HH53 pKa = 6.2KK54 pKa = 10.47GGQHH58 pKa = 4.35KK59 pKa = 9.92RR60 pKa = 11.84YY61 pKa = 10.13YY62 pKa = 10.45RR63 pKa = 11.84IIDD66 pKa = 3.88FKK68 pKa = 11.13RR69 pKa = 11.84NEE71 pKa = 4.09YY72 pKa = 10.0GIKK75 pKa = 10.4GYY77 pKa = 8.94VKK79 pKa = 10.26RR80 pKa = 11.84IEE82 pKa = 3.97YY83 pKa = 10.47DD84 pKa = 3.15PNRR87 pKa = 11.84NCNIALIYY95 pKa = 9.37YY96 pKa = 10.35LNGKK100 pKa = 8.7KK101 pKa = 10.07KK102 pKa = 10.93YY103 pKa = 9.63ILHH106 pKa = 6.7PKK108 pKa = 8.87NLNINDD114 pKa = 4.36EE115 pKa = 4.52IISNYY120 pKa = 7.41KK121 pKa = 9.2TPIKK125 pKa = 10.23IGNNLLIKK133 pKa = 10.11NIPLGTLIHH142 pKa = 6.44CLEE145 pKa = 4.56LLPFKK150 pKa = 10.45GAQIARR156 pKa = 11.84SAGSSALIISRR167 pKa = 11.84EE168 pKa = 3.78KK169 pKa = 10.65KK170 pKa = 8.08YY171 pKa = 8.72TQVRR175 pKa = 11.84LKK177 pKa = 10.6SNEE180 pKa = 3.02IRR182 pKa = 11.84RR183 pKa = 11.84INNNCFATIGEE194 pKa = 4.4IGNEE198 pKa = 3.59KK199 pKa = 10.58HH200 pKa = 6.83NLIKK204 pKa = 10.6LGKK207 pKa = 10.11AGIKK211 pKa = 9.83RR212 pKa = 11.84NLGIRR217 pKa = 11.84PTVRR221 pKa = 11.84GVAMNPIDD229 pKa = 4.06HH230 pKa = 6.89PHH232 pKa = 6.54GGGEE236 pKa = 4.02GRR238 pKa = 11.84TSGGRR243 pKa = 11.84HH244 pKa = 5.87PVTPWGKK251 pKa = 8.02KK252 pKa = 6.7TKK254 pKa = 10.06GKK256 pKa = 8.43KK257 pKa = 7.59TRR259 pKa = 11.84NKK261 pKa = 10.49KK262 pKa = 10.26KK263 pKa = 9.82ISNNMIINNKK273 pKa = 7.88NDD275 pKa = 3.56KK276 pKa = 10.67IFF278 pKa = 3.77
Molecular weight: 31.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
62563 |
37 |
1291 |
303.7 |
36.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.131 | 1.073 ± 0.051 |
2.267 ± 0.09 | 3.277 ± 0.142 |
6.923 ± 0.251 | 3.697 ± 0.178 |
0.964 ± 0.046 | 17.49 ± 0.258 |
17.948 ± 0.349 | 9.296 ± 0.148 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.232 ± 0.062 | 12.813 ± 0.233 |
1.966 ± 0.078 | 1.181 ± 0.061 |
2.036 ± 0.09 | 5.185 ± 0.144 |
2.612 ± 0.111 | 2.048 ± 0.117 |
0.599 ± 0.052 | 5.748 ± 0.133 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
