
Goose hemorrhagic polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Gammapolyomavirus; Anser anser polyomavirus 1
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
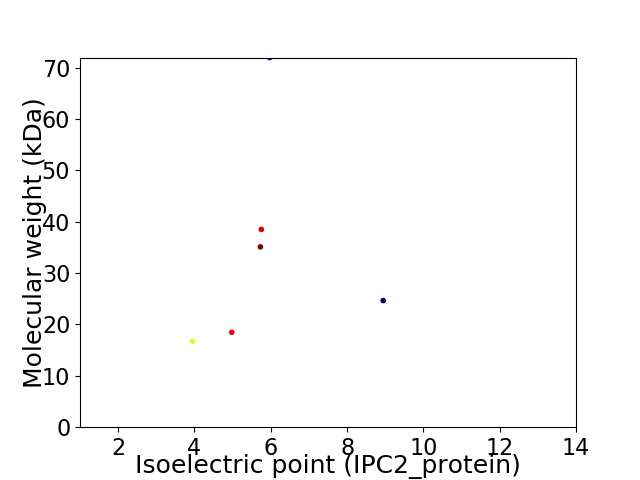
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

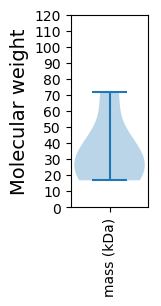
Protein with the lowest isoelectric point:
>tr|Q80FI6|Q80FI6_9POLY ORF-X OS=Goose hemorrhagic polyomavirus OX=208491 GN=ORF-X PE=4 SV=1
MM1 pKa = 7.58SEE3 pKa = 4.07ATRR6 pKa = 11.84KK7 pKa = 9.97PMDD10 pKa = 3.6SEE12 pKa = 4.69GPRR15 pKa = 11.84AASAAAAPRR24 pKa = 11.84TEE26 pKa = 4.8DD27 pKa = 3.91SEE29 pKa = 4.82GPPSAAGGSVGDD41 pKa = 4.24IPAPVPASPAPEE53 pKa = 4.16EE54 pKa = 4.28ASGVPPVPEE63 pKa = 3.75KK64 pKa = 10.88AGPDD68 pKa = 3.54GAAAPHH74 pKa = 6.23ATAGPAGTSSTAAEE88 pKa = 4.33VTEE91 pKa = 5.12DD92 pKa = 4.18PPEE95 pKa = 4.38STPSPEE101 pKa = 4.06RR102 pKa = 11.84DD103 pKa = 3.09GVSPKK108 pKa = 10.72EE109 pKa = 4.25PLTQPSTGSSSTDD122 pKa = 3.21LSPPDD127 pKa = 4.19SPHH130 pKa = 7.64ASDD133 pKa = 6.02DD134 pKa = 4.4DD135 pKa = 4.17MDD137 pKa = 3.91TSPYY141 pKa = 10.59YY142 pKa = 10.31EE143 pKa = 4.81GSCTCTFSLKK153 pKa = 10.04VCCNICSGCNEE164 pKa = 4.56FLNSDD169 pKa = 3.4
MM1 pKa = 7.58SEE3 pKa = 4.07ATRR6 pKa = 11.84KK7 pKa = 9.97PMDD10 pKa = 3.6SEE12 pKa = 4.69GPRR15 pKa = 11.84AASAAAAPRR24 pKa = 11.84TEE26 pKa = 4.8DD27 pKa = 3.91SEE29 pKa = 4.82GPPSAAGGSVGDD41 pKa = 4.24IPAPVPASPAPEE53 pKa = 4.16EE54 pKa = 4.28ASGVPPVPEE63 pKa = 3.75KK64 pKa = 10.88AGPDD68 pKa = 3.54GAAAPHH74 pKa = 6.23ATAGPAGTSSTAAEE88 pKa = 4.33VTEE91 pKa = 5.12DD92 pKa = 4.18PPEE95 pKa = 4.38STPSPEE101 pKa = 4.06RR102 pKa = 11.84DD103 pKa = 3.09GVSPKK108 pKa = 10.72EE109 pKa = 4.25PLTQPSTGSSSTDD122 pKa = 3.21LSPPDD127 pKa = 4.19SPHH130 pKa = 7.64ASDD133 pKa = 6.02DD134 pKa = 4.4DD135 pKa = 4.17MDD137 pKa = 3.91TSPYY141 pKa = 10.59YY142 pKa = 10.31EE143 pKa = 4.81GSCTCTFSLKK153 pKa = 10.04VCCNICSGCNEE164 pKa = 4.56FLNSDD169 pKa = 3.4
Molecular weight: 16.76 kDa
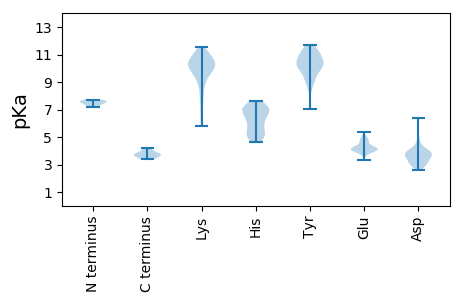
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q80FI5|Q80FI5_9POLY Minor capsid protein OS=Goose hemorrhagic polyomavirus OX=208491 GN=VP2 PE=3 SV=1
MM1 pKa = 7.72AIVEE5 pKa = 3.99WRR7 pKa = 11.84PYY9 pKa = 10.58VDD11 pKa = 3.42YY12 pKa = 10.93LFPGFEE18 pKa = 4.04TFVNQFYY25 pKa = 10.19WINPLEE31 pKa = 4.27WGPTLFQRR39 pKa = 11.84VGQAFWDD46 pKa = 3.83LLITEE51 pKa = 4.71GRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84LGEE58 pKa = 4.0ATSAAAQTSAQYY70 pKa = 11.06VYY72 pKa = 11.45DD73 pKa = 4.9LIARR77 pKa = 11.84TAEE80 pKa = 3.98RR81 pKa = 11.84ARR83 pKa = 11.84WTVVEE88 pKa = 4.77GPGEE92 pKa = 4.14AYY94 pKa = 10.24RR95 pKa = 11.84GLIEE99 pKa = 4.74YY100 pKa = 10.39YY101 pKa = 10.99GEE103 pKa = 4.18LPPWKK108 pKa = 9.18PHH110 pKa = 4.93SLHH113 pKa = 7.13RR114 pKa = 11.84GLPWEE119 pKa = 4.26QKK121 pKa = 5.82TTEE124 pKa = 4.15TGGNGQPRR132 pKa = 11.84QQPDD136 pKa = 3.01SGEE139 pKa = 4.1TVYY142 pKa = 10.46QQPVPGGANQRR153 pKa = 11.84AAPDD157 pKa = 3.11WMLPLILGLYY167 pKa = 7.05GTVYY171 pKa = 10.26PGWKK175 pKa = 10.19AEE177 pKa = 3.95VALLEE182 pKa = 4.9KK183 pKa = 10.69EE184 pKa = 4.99DD185 pKa = 3.77GSQGKK190 pKa = 7.81KK191 pKa = 6.51TKK193 pKa = 10.14KK194 pKa = 8.65RR195 pKa = 11.84TRR197 pKa = 11.84ASPQTSGKK205 pKa = 9.48RR206 pKa = 11.84RR207 pKa = 11.84YY208 pKa = 9.61RR209 pKa = 11.84GPRR212 pKa = 11.84GQDD215 pKa = 2.64RR216 pKa = 11.84AA217 pKa = 3.8
MM1 pKa = 7.72AIVEE5 pKa = 3.99WRR7 pKa = 11.84PYY9 pKa = 10.58VDD11 pKa = 3.42YY12 pKa = 10.93LFPGFEE18 pKa = 4.04TFVNQFYY25 pKa = 10.19WINPLEE31 pKa = 4.27WGPTLFQRR39 pKa = 11.84VGQAFWDD46 pKa = 3.83LLITEE51 pKa = 4.71GRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84LGEE58 pKa = 4.0ATSAAAQTSAQYY70 pKa = 11.06VYY72 pKa = 11.45DD73 pKa = 4.9LIARR77 pKa = 11.84TAEE80 pKa = 3.98RR81 pKa = 11.84ARR83 pKa = 11.84WTVVEE88 pKa = 4.77GPGEE92 pKa = 4.14AYY94 pKa = 10.24RR95 pKa = 11.84GLIEE99 pKa = 4.74YY100 pKa = 10.39YY101 pKa = 10.99GEE103 pKa = 4.18LPPWKK108 pKa = 9.18PHH110 pKa = 4.93SLHH113 pKa = 7.13RR114 pKa = 11.84GLPWEE119 pKa = 4.26QKK121 pKa = 5.82TTEE124 pKa = 4.15TGGNGQPRR132 pKa = 11.84QQPDD136 pKa = 3.01SGEE139 pKa = 4.1TVYY142 pKa = 10.46QQPVPGGANQRR153 pKa = 11.84AAPDD157 pKa = 3.11WMLPLILGLYY167 pKa = 7.05GTVYY171 pKa = 10.26PGWKK175 pKa = 10.19AEE177 pKa = 3.95VALLEE182 pKa = 4.9KK183 pKa = 10.69EE184 pKa = 4.99DD185 pKa = 3.77GSQGKK190 pKa = 7.81KK191 pKa = 6.51TKK193 pKa = 10.14KK194 pKa = 8.65RR195 pKa = 11.84TRR197 pKa = 11.84ASPQTSGKK205 pKa = 9.48RR206 pKa = 11.84RR207 pKa = 11.84YY208 pKa = 9.61RR209 pKa = 11.84GPRR212 pKa = 11.84GQDD215 pKa = 2.64RR216 pKa = 11.84AA217 pKa = 3.8
Molecular weight: 24.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1861 |
160 |
636 |
310.2 |
34.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.598 ± 1.713 | 2.149 ± 0.588 |
4.782 ± 0.658 | 7.792 ± 0.653 |
3.385 ± 0.472 | 7.899 ± 1.18 |
1.934 ± 0.363 | 3.815 ± 0.771 |
4.89 ± 0.808 | 8.866 ± 0.994 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.203 ± 0.387 | 3.332 ± 0.632 |
7.577 ± 1.4 | 4.03 ± 0.57 |
5.75 ± 0.541 | 6.663 ± 0.92 |
5.75 ± 0.581 | 5.75 ± 0.933 |
1.343 ± 0.514 | 3.493 ± 0.381 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
