
Malassezia sympodialis (strain ATCC 42132) (Atopic eczema-associated yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Malasseziomycetes; Malasseziales; Malasseziaceae; Malassezia; Malassezia sympodialis
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
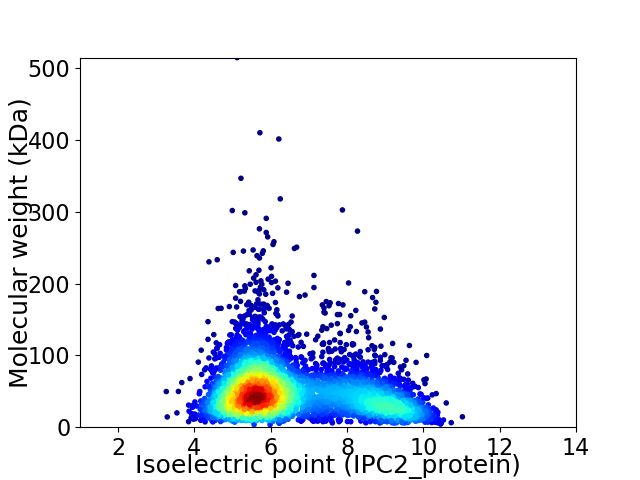
Virtual 2D-PAGE plot for 4501 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M8A3B6|A0A1M8A3B6_MALS4 Uncharacterized protein OS=Malassezia sympodialis (strain ATCC 42132) OX=1230383 GN=MSYG_1299 PE=4 SV=1
MM1 pKa = 7.3AVSYY5 pKa = 10.71YY6 pKa = 10.93DD7 pKa = 4.0EE8 pKa = 6.04IEE10 pKa = 4.46LEE12 pKa = 4.21DD13 pKa = 3.59MEE15 pKa = 5.19FDD17 pKa = 3.31EE18 pKa = 5.77GKK20 pKa = 9.97QAFTYY25 pKa = 8.74PCPCGDD31 pKa = 3.36KK32 pKa = 11.11FEE34 pKa = 5.17ISLEE38 pKa = 3.95QLKK41 pKa = 10.75NEE43 pKa = 4.14EE44 pKa = 4.81DD45 pKa = 3.37IARR48 pKa = 11.84CPSCTLLIRR57 pKa = 11.84VIYY60 pKa = 10.43DD61 pKa = 3.48PLDD64 pKa = 3.6FDD66 pKa = 4.39
MM1 pKa = 7.3AVSYY5 pKa = 10.71YY6 pKa = 10.93DD7 pKa = 4.0EE8 pKa = 6.04IEE10 pKa = 4.46LEE12 pKa = 4.21DD13 pKa = 3.59MEE15 pKa = 5.19FDD17 pKa = 3.31EE18 pKa = 5.77GKK20 pKa = 9.97QAFTYY25 pKa = 8.74PCPCGDD31 pKa = 3.36KK32 pKa = 11.11FEE34 pKa = 5.17ISLEE38 pKa = 3.95QLKK41 pKa = 10.75NEE43 pKa = 4.14EE44 pKa = 4.81DD45 pKa = 3.37IARR48 pKa = 11.84CPSCTLLIRR57 pKa = 11.84VIYY60 pKa = 10.43DD61 pKa = 3.48PLDD64 pKa = 3.6FDD66 pKa = 4.39
Molecular weight: 7.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M8A1W2|A0A1M8A1W2_MALS4 Similar to S.cerevisiae protein RCF1 (Cytochrome c oxidase subunit) OS=Malassezia sympodialis (strain ATCC 42132) OX=1230383 GN=MSYG_0519 PE=4 SV=1
MM1 pKa = 6.93NTPGLVAVAQAFVRR15 pKa = 11.84PGVLVPHH22 pKa = 6.46LQVASLRR29 pKa = 11.84LIDD32 pKa = 3.63WRR34 pKa = 11.84QVRR37 pKa = 11.84ASGVRR42 pKa = 11.84YY43 pKa = 9.43IVYY46 pKa = 10.61DD47 pKa = 3.61KK48 pKa = 11.04DD49 pKa = 3.46NCLTRR54 pKa = 11.84PHH56 pKa = 6.97HH57 pKa = 7.08DD58 pKa = 3.28ALAPALEE65 pKa = 4.39ATWDD69 pKa = 3.29EE70 pKa = 4.52CKK72 pKa = 10.68AVFGEE77 pKa = 4.27EE78 pKa = 3.63NLLIVSNSAGSSQDD92 pKa = 3.27PTGLAAEE99 pKa = 4.29QVSSKK104 pKa = 10.84LGVPILCHH112 pKa = 5.86AVKK115 pKa = 10.62KK116 pKa = 10.2PGRR119 pKa = 11.84ACAQQVVDD127 pKa = 4.56YY128 pKa = 10.71VASQHH133 pKa = 6.28LRR135 pKa = 11.84SPDD138 pKa = 3.23AAHH141 pKa = 7.26PPRR144 pKa = 11.84LLVVGDD150 pKa = 5.1RR151 pKa = 11.84ITTDD155 pKa = 2.75MAFSYY160 pKa = 10.49RR161 pKa = 11.84IASLLGRR168 pKa = 11.84AYY170 pKa = 9.51PHH172 pKa = 6.72EE173 pKa = 5.58RR174 pKa = 11.84DD175 pKa = 3.28LCIGVLTHH183 pKa = 6.24EE184 pKa = 4.11LWGRR188 pKa = 11.84EE189 pKa = 3.93QLGTRR194 pKa = 11.84VMRR197 pKa = 11.84MLEE200 pKa = 4.09NTVLRR205 pKa = 11.84QLVRR209 pKa = 11.84VGIPPGGTWTARR221 pKa = 11.84GAPSPVCSAWVRR233 pKa = 11.84AVAPADD239 pKa = 3.87AAPLPAPRR247 pKa = 11.84KK248 pKa = 9.82ARR250 pKa = 11.84THH252 pKa = 5.25MRR254 pKa = 11.84WLAPLVDD261 pKa = 3.74AWRR264 pKa = 11.84TIVRR268 pKa = 11.84EE269 pKa = 3.96TFGSVRR275 pKa = 11.84RR276 pKa = 11.84VRR278 pKa = 11.84EE279 pKa = 4.24LTWSVCTNAPTRR291 pKa = 11.84SWRR294 pKa = 11.84STWRR298 pKa = 11.84KK299 pKa = 7.09PQRR302 pKa = 11.84PPPAPWSRR310 pKa = 11.84SMSTSARR317 pKa = 11.84ARR319 pKa = 11.84GALHH323 pKa = 6.81EE324 pKa = 4.56KK325 pKa = 9.54PVPRR329 pKa = 11.84RR330 pKa = 11.84VPTADD335 pKa = 3.16APRR338 pKa = 11.84APRR341 pKa = 11.84AVPTWLGIPRR351 pKa = 11.84IRR353 pKa = 11.84WLMALATLVLLPLGFMGGMKK373 pKa = 10.37LSEE376 pKa = 4.11LVEE379 pKa = 3.93RR380 pKa = 11.84WRR382 pKa = 11.84IGDD385 pKa = 3.8LSHH388 pKa = 6.86EE389 pKa = 4.43GEE391 pKa = 4.58ARR393 pKa = 11.84VDD395 pKa = 3.74APPPAPAPEE404 pKa = 4.67PDD406 pKa = 3.44VEE408 pKa = 4.33EE409 pKa = 4.3TQVQLRR415 pKa = 11.84KK416 pKa = 10.03KK417 pKa = 9.97ISSLEE422 pKa = 3.94LEE424 pKa = 4.54HH425 pKa = 6.42FHH427 pKa = 6.8LRR429 pKa = 11.84RR430 pKa = 11.84EE431 pKa = 4.01RR432 pKa = 11.84SVLDD436 pKa = 3.83DD437 pKa = 3.5KK438 pKa = 11.2LARR441 pKa = 11.84LAARR445 pKa = 11.84PAA447 pKa = 3.4
MM1 pKa = 6.93NTPGLVAVAQAFVRR15 pKa = 11.84PGVLVPHH22 pKa = 6.46LQVASLRR29 pKa = 11.84LIDD32 pKa = 3.63WRR34 pKa = 11.84QVRR37 pKa = 11.84ASGVRR42 pKa = 11.84YY43 pKa = 9.43IVYY46 pKa = 10.61DD47 pKa = 3.61KK48 pKa = 11.04DD49 pKa = 3.46NCLTRR54 pKa = 11.84PHH56 pKa = 6.97HH57 pKa = 7.08DD58 pKa = 3.28ALAPALEE65 pKa = 4.39ATWDD69 pKa = 3.29EE70 pKa = 4.52CKK72 pKa = 10.68AVFGEE77 pKa = 4.27EE78 pKa = 3.63NLLIVSNSAGSSQDD92 pKa = 3.27PTGLAAEE99 pKa = 4.29QVSSKK104 pKa = 10.84LGVPILCHH112 pKa = 5.86AVKK115 pKa = 10.62KK116 pKa = 10.2PGRR119 pKa = 11.84ACAQQVVDD127 pKa = 4.56YY128 pKa = 10.71VASQHH133 pKa = 6.28LRR135 pKa = 11.84SPDD138 pKa = 3.23AAHH141 pKa = 7.26PPRR144 pKa = 11.84LLVVGDD150 pKa = 5.1RR151 pKa = 11.84ITTDD155 pKa = 2.75MAFSYY160 pKa = 10.49RR161 pKa = 11.84IASLLGRR168 pKa = 11.84AYY170 pKa = 9.51PHH172 pKa = 6.72EE173 pKa = 5.58RR174 pKa = 11.84DD175 pKa = 3.28LCIGVLTHH183 pKa = 6.24EE184 pKa = 4.11LWGRR188 pKa = 11.84EE189 pKa = 3.93QLGTRR194 pKa = 11.84VMRR197 pKa = 11.84MLEE200 pKa = 4.09NTVLRR205 pKa = 11.84QLVRR209 pKa = 11.84VGIPPGGTWTARR221 pKa = 11.84GAPSPVCSAWVRR233 pKa = 11.84AVAPADD239 pKa = 3.87AAPLPAPRR247 pKa = 11.84KK248 pKa = 9.82ARR250 pKa = 11.84THH252 pKa = 5.25MRR254 pKa = 11.84WLAPLVDD261 pKa = 3.74AWRR264 pKa = 11.84TIVRR268 pKa = 11.84EE269 pKa = 3.96TFGSVRR275 pKa = 11.84RR276 pKa = 11.84VRR278 pKa = 11.84EE279 pKa = 4.24LTWSVCTNAPTRR291 pKa = 11.84SWRR294 pKa = 11.84STWRR298 pKa = 11.84KK299 pKa = 7.09PQRR302 pKa = 11.84PPPAPWSRR310 pKa = 11.84SMSTSARR317 pKa = 11.84ARR319 pKa = 11.84GALHH323 pKa = 6.81EE324 pKa = 4.56KK325 pKa = 9.54PVPRR329 pKa = 11.84RR330 pKa = 11.84VPTADD335 pKa = 3.16APRR338 pKa = 11.84APRR341 pKa = 11.84AVPTWLGIPRR351 pKa = 11.84IRR353 pKa = 11.84WLMALATLVLLPLGFMGGMKK373 pKa = 10.37LSEE376 pKa = 4.11LVEE379 pKa = 3.93RR380 pKa = 11.84WRR382 pKa = 11.84IGDD385 pKa = 3.8LSHH388 pKa = 6.86EE389 pKa = 4.43GEE391 pKa = 4.58ARR393 pKa = 11.84VDD395 pKa = 3.74APPPAPAPEE404 pKa = 4.67PDD406 pKa = 3.44VEE408 pKa = 4.33EE409 pKa = 4.3TQVQLRR415 pKa = 11.84KK416 pKa = 10.03KK417 pKa = 9.97ISSLEE422 pKa = 3.94LEE424 pKa = 4.54HH425 pKa = 6.42FHH427 pKa = 6.8LRR429 pKa = 11.84RR430 pKa = 11.84EE431 pKa = 4.01RR432 pKa = 11.84SVLDD436 pKa = 3.83DD437 pKa = 3.5KK438 pKa = 11.2LARR441 pKa = 11.84LAARR445 pKa = 11.84PAA447 pKa = 3.4
Molecular weight: 49.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2239772 |
34 |
4822 |
497.6 |
54.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.956 ± 0.08 | 1.226 ± 0.013 |
5.667 ± 0.026 | 6.03 ± 0.033 |
3.15 ± 0.021 | 6.107 ± 0.032 |
2.936 ± 0.02 | 3.62 ± 0.028 |
3.536 ± 0.037 | 9.986 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.015 | 2.585 ± 0.027 |
6.601 ± 0.042 | 4.214 ± 0.021 |
7.114 ± 0.034 | 7.48 ± 0.043 |
5.523 ± 0.02 | 6.766 ± 0.025 |
1.459 ± 0.013 | 2.538 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
