
Pigeonpea sterility mosaic emaravirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
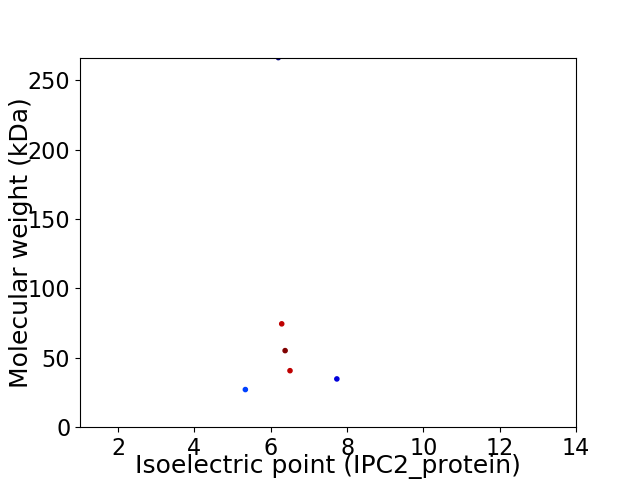
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
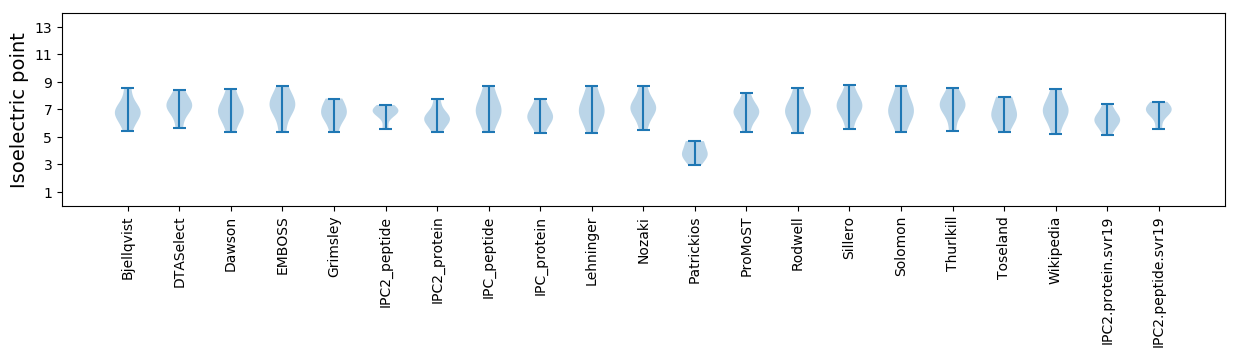
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0G4DI13|A0A0G4DI13_9VIRU p5 protein OS=Pigeonpea sterility mosaic emaravirus 2 OX=1980430 GN=p5 PE=4 SV=1
MM1 pKa = 7.54ASKK4 pKa = 10.67GFEE7 pKa = 3.76RR8 pKa = 11.84TFNICNTANLEE19 pKa = 4.08EE20 pKa = 5.13LYY22 pKa = 10.91SEE24 pKa = 5.47LEE26 pKa = 3.94SQLYY30 pKa = 9.65YY31 pKa = 10.89SAIEE35 pKa = 3.98YY36 pKa = 8.54MLSIDD41 pKa = 4.5LYY43 pKa = 11.47GEE45 pKa = 3.82IAVLRR50 pKa = 11.84EE51 pKa = 3.76IYY53 pKa = 10.25EE54 pKa = 4.44DD55 pKa = 3.54NSLDD59 pKa = 3.75LSILISSAPYY69 pKa = 10.38LIIEE73 pKa = 4.55GNSVSRR79 pKa = 11.84KK80 pKa = 6.33SVKK83 pKa = 10.3YY84 pKa = 6.24FTRR87 pKa = 11.84YY88 pKa = 9.46LQFSMSLFGVSEE100 pKa = 4.27DD101 pKa = 3.97ACSGLCPDD109 pKa = 3.27YY110 pKa = 11.05HH111 pKa = 8.48IFGSVHH117 pKa = 5.46RR118 pKa = 11.84QKK120 pKa = 10.89QILPYY125 pKa = 9.99IPNNSKK131 pKa = 10.69YY132 pKa = 10.73LLVDD136 pKa = 3.11TSILKK141 pKa = 10.09EE142 pKa = 4.33SVWDD146 pKa = 3.5VMRR149 pKa = 11.84GVDD152 pKa = 3.64PYY154 pKa = 11.34KK155 pKa = 10.5VLEE158 pKa = 4.16RR159 pKa = 11.84VYY161 pKa = 10.85LDD163 pKa = 3.33QPRR166 pKa = 11.84SMDD169 pKa = 3.47LSEE172 pKa = 4.21STVAFNFHH180 pKa = 6.14IMAISYY186 pKa = 10.22ISIRR190 pKa = 11.84SSSKK194 pKa = 10.96LKK196 pKa = 10.5IEE198 pKa = 4.22CKK200 pKa = 10.09KK201 pKa = 10.61LRR203 pKa = 11.84LSSRR207 pKa = 11.84IFSDD211 pKa = 3.05NGATSASHH219 pKa = 6.75VKK221 pKa = 10.31DD222 pKa = 3.82AEE224 pKa = 4.22FNSIDD229 pKa = 3.89KK230 pKa = 10.68SHH232 pKa = 6.81SPSQLEE238 pKa = 3.9
MM1 pKa = 7.54ASKK4 pKa = 10.67GFEE7 pKa = 3.76RR8 pKa = 11.84TFNICNTANLEE19 pKa = 4.08EE20 pKa = 5.13LYY22 pKa = 10.91SEE24 pKa = 5.47LEE26 pKa = 3.94SQLYY30 pKa = 9.65YY31 pKa = 10.89SAIEE35 pKa = 3.98YY36 pKa = 8.54MLSIDD41 pKa = 4.5LYY43 pKa = 11.47GEE45 pKa = 3.82IAVLRR50 pKa = 11.84EE51 pKa = 3.76IYY53 pKa = 10.25EE54 pKa = 4.44DD55 pKa = 3.54NSLDD59 pKa = 3.75LSILISSAPYY69 pKa = 10.38LIIEE73 pKa = 4.55GNSVSRR79 pKa = 11.84KK80 pKa = 6.33SVKK83 pKa = 10.3YY84 pKa = 6.24FTRR87 pKa = 11.84YY88 pKa = 9.46LQFSMSLFGVSEE100 pKa = 4.27DD101 pKa = 3.97ACSGLCPDD109 pKa = 3.27YY110 pKa = 11.05HH111 pKa = 8.48IFGSVHH117 pKa = 5.46RR118 pKa = 11.84QKK120 pKa = 10.89QILPYY125 pKa = 9.99IPNNSKK131 pKa = 10.69YY132 pKa = 10.73LLVDD136 pKa = 3.11TSILKK141 pKa = 10.09EE142 pKa = 4.33SVWDD146 pKa = 3.5VMRR149 pKa = 11.84GVDD152 pKa = 3.64PYY154 pKa = 11.34KK155 pKa = 10.5VLEE158 pKa = 4.16RR159 pKa = 11.84VYY161 pKa = 10.85LDD163 pKa = 3.33QPRR166 pKa = 11.84SMDD169 pKa = 3.47LSEE172 pKa = 4.21STVAFNFHH180 pKa = 6.14IMAISYY186 pKa = 10.22ISIRR190 pKa = 11.84SSSKK194 pKa = 10.96LKK196 pKa = 10.5IEE198 pKa = 4.22CKK200 pKa = 10.09KK201 pKa = 10.61LRR203 pKa = 11.84LSSRR207 pKa = 11.84IFSDD211 pKa = 3.05NGATSASHH219 pKa = 6.75VKK221 pKa = 10.31DD222 pKa = 3.82AEE224 pKa = 4.22FNSIDD229 pKa = 3.89KK230 pKa = 10.68SHH232 pKa = 6.81SPSQLEE238 pKa = 3.9
Molecular weight: 27.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G4DD52|A0A0G4DD52_9VIRU Movment protein OS=Pigeonpea sterility mosaic emaravirus 2 OX=1980430 GN=putative movement protein PE=4 SV=1
MM1 pKa = 7.45PPKK4 pKa = 10.23RR5 pKa = 11.84SISSTSAPKK14 pKa = 10.06AQPGALQEE22 pKa = 3.97NSILLGSGSKK32 pKa = 10.52LRR34 pKa = 11.84VIKK37 pKa = 9.15LTNVVNGKK45 pKa = 8.36LTNAEE50 pKa = 4.14SSEE53 pKa = 4.15LEE55 pKa = 4.59PIDD58 pKa = 4.89VEE60 pKa = 4.35VSPFTSTSQNVSNFTLHH77 pKa = 6.94NYY79 pKa = 9.59RR80 pKa = 11.84SCCHH84 pKa = 6.39VDD86 pKa = 3.49TVAAHH91 pKa = 6.49LSKK94 pKa = 10.94SKK96 pKa = 10.35EE97 pKa = 3.98IKK99 pKa = 10.24EE100 pKa = 4.08KK101 pKa = 10.49LQSRR105 pKa = 11.84NLKK108 pKa = 10.18LATSKK113 pKa = 10.92DD114 pKa = 3.37DD115 pKa = 3.57HH116 pKa = 7.16LVVVKK121 pKa = 10.64DD122 pKa = 3.93MNDD125 pKa = 3.05STVINVVSFNKK136 pKa = 10.12ACAIMAAGILKK147 pKa = 8.91HH148 pKa = 5.84TFDD151 pKa = 5.8EE152 pKa = 4.45EE153 pKa = 4.4FDD155 pKa = 3.74WKK157 pKa = 8.97TGKK160 pKa = 9.66YY161 pKa = 10.17VKK163 pKa = 9.57TDD165 pKa = 2.99NKK167 pKa = 9.98TKK169 pKa = 10.43VVPNPAIINRR179 pKa = 11.84LAGQMGLSAGNPYY192 pKa = 10.72YY193 pKa = 10.44WMIVPGYY200 pKa = 8.6EE201 pKa = 4.21FLYY204 pKa = 10.04EE205 pKa = 4.26LYY207 pKa = 9.64PAEE210 pKa = 4.09VLAYY214 pKa = 9.16TLVRR218 pKa = 11.84LQFRR222 pKa = 11.84KK223 pKa = 10.2NLNIPDD229 pKa = 4.13KK230 pKa = 9.49MTDD233 pKa = 3.1ADD235 pKa = 3.86IVSSLVMKK243 pKa = 9.43MNRR246 pKa = 11.84IHH248 pKa = 7.37KK249 pKa = 10.08LEE251 pKa = 3.91QTSFDD256 pKa = 4.03DD257 pKa = 5.13AINTIGRR264 pKa = 11.84DD265 pKa = 3.81NVSEE269 pKa = 4.56AYY271 pKa = 10.51VEE273 pKa = 3.99LARR276 pKa = 11.84DD277 pKa = 3.33IGSTSKK283 pKa = 9.71TKK285 pKa = 10.82RR286 pKa = 11.84NDD288 pKa = 2.92EE289 pKa = 4.45AIIKK293 pKa = 9.57FKK295 pKa = 10.93EE296 pKa = 4.42LISTFLPALEE306 pKa = 4.26ADD308 pKa = 4.18RR309 pKa = 11.84LASLL313 pKa = 4.46
MM1 pKa = 7.45PPKK4 pKa = 10.23RR5 pKa = 11.84SISSTSAPKK14 pKa = 10.06AQPGALQEE22 pKa = 3.97NSILLGSGSKK32 pKa = 10.52LRR34 pKa = 11.84VIKK37 pKa = 9.15LTNVVNGKK45 pKa = 8.36LTNAEE50 pKa = 4.14SSEE53 pKa = 4.15LEE55 pKa = 4.59PIDD58 pKa = 4.89VEE60 pKa = 4.35VSPFTSTSQNVSNFTLHH77 pKa = 6.94NYY79 pKa = 9.59RR80 pKa = 11.84SCCHH84 pKa = 6.39VDD86 pKa = 3.49TVAAHH91 pKa = 6.49LSKK94 pKa = 10.94SKK96 pKa = 10.35EE97 pKa = 3.98IKK99 pKa = 10.24EE100 pKa = 4.08KK101 pKa = 10.49LQSRR105 pKa = 11.84NLKK108 pKa = 10.18LATSKK113 pKa = 10.92DD114 pKa = 3.37DD115 pKa = 3.57HH116 pKa = 7.16LVVVKK121 pKa = 10.64DD122 pKa = 3.93MNDD125 pKa = 3.05STVINVVSFNKK136 pKa = 10.12ACAIMAAGILKK147 pKa = 8.91HH148 pKa = 5.84TFDD151 pKa = 5.8EE152 pKa = 4.45EE153 pKa = 4.4FDD155 pKa = 3.74WKK157 pKa = 8.97TGKK160 pKa = 9.66YY161 pKa = 10.17VKK163 pKa = 9.57TDD165 pKa = 2.99NKK167 pKa = 9.98TKK169 pKa = 10.43VVPNPAIINRR179 pKa = 11.84LAGQMGLSAGNPYY192 pKa = 10.72YY193 pKa = 10.44WMIVPGYY200 pKa = 8.6EE201 pKa = 4.21FLYY204 pKa = 10.04EE205 pKa = 4.26LYY207 pKa = 9.64PAEE210 pKa = 4.09VLAYY214 pKa = 9.16TLVRR218 pKa = 11.84LQFRR222 pKa = 11.84KK223 pKa = 10.2NLNIPDD229 pKa = 4.13KK230 pKa = 9.49MTDD233 pKa = 3.1ADD235 pKa = 3.86IVSSLVMKK243 pKa = 9.43MNRR246 pKa = 11.84IHH248 pKa = 7.37KK249 pKa = 10.08LEE251 pKa = 3.91QTSFDD256 pKa = 4.03DD257 pKa = 5.13AINTIGRR264 pKa = 11.84DD265 pKa = 3.81NVSEE269 pKa = 4.56AYY271 pKa = 10.51VEE273 pKa = 3.99LARR276 pKa = 11.84DD277 pKa = 3.33IGSTSKK283 pKa = 9.71TKK285 pKa = 10.82RR286 pKa = 11.84NDD288 pKa = 2.92EE289 pKa = 4.45AIIKK293 pKa = 9.57FKK295 pKa = 10.93EE296 pKa = 4.42LISTFLPALEE306 pKa = 4.26ADD308 pKa = 4.18RR309 pKa = 11.84LASLL313 pKa = 4.46
Molecular weight: 34.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4328 |
238 |
2294 |
721.3 |
83.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.043 ± 0.59 | 1.941 ± 0.398 |
6.285 ± 0.409 | 5.799 ± 0.417 |
4.321 ± 0.209 | 3.581 ± 0.387 |
2.518 ± 0.285 | 9.219 ± 0.381 |
7.971 ± 0.234 | 9.011 ± 0.737 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.286 | 7.093 ± 0.468 |
3.189 ± 0.181 | 2.518 ± 0.33 |
3.697 ± 0.207 | 8.272 ± 0.741 |
5.869 ± 0.333 | 5.73 ± 0.774 |
0.439 ± 0.128 | 5.869 ± 0.489 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
