
Ustilaginoidea virens unassigned RNA virus HNND-1
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
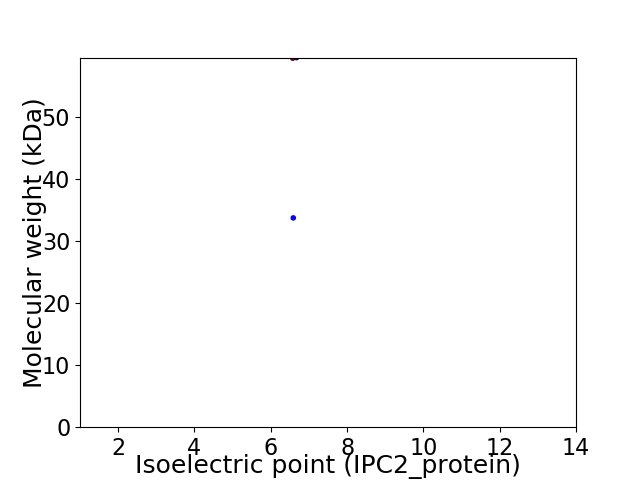
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3ZCM4|A0A0G3ZCM4_9VIRU Uncharacterized protein OS=Ustilaginoidea virens unassigned RNA virus HNND-1 OX=1670975 PE=4 SV=1
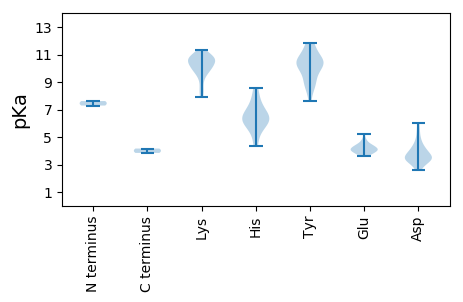
MM1 pKa = 7.29TCMAGPSEE9 pKa = 4.35EE10 pKa = 4.17VVMSEE15 pKa = 4.29FVPPAISDD23 pKa = 4.07LYY25 pKa = 11.28KK26 pKa = 10.89HH27 pKa = 6.0MEE29 pKa = 4.12GFGNRR34 pKa = 11.84NPGRR38 pKa = 11.84VKK40 pKa = 9.91PTTDD44 pKa = 2.67TYY46 pKa = 11.75AGLFSEE52 pKa = 4.69LRR54 pKa = 11.84AVEE57 pKa = 3.93GWTRR61 pKa = 11.84SISGWLDD68 pKa = 3.06PALLPKK74 pKa = 10.11IVVPSQTSPGIRR86 pKa = 11.84WKK88 pKa = 10.76RR89 pKa = 11.84LGYY92 pKa = 7.65RR93 pKa = 11.84TKK95 pKa = 10.6RR96 pKa = 11.84AALMPAVVEE105 pKa = 4.05ASRR108 pKa = 11.84LVRR111 pKa = 11.84RR112 pKa = 11.84MADD115 pKa = 2.77TGEE118 pKa = 4.23KK119 pKa = 10.75YY120 pKa = 9.59EE121 pKa = 4.06VPPAGVAGRR130 pKa = 11.84GKK132 pKa = 10.54RR133 pKa = 11.84LDD135 pKa = 3.42SCRR138 pKa = 11.84DD139 pKa = 3.1TNAPDD144 pKa = 3.4RR145 pKa = 11.84KK146 pKa = 9.02EE147 pKa = 3.67GRR149 pKa = 11.84LIVMPDD155 pKa = 3.6LVRR158 pKa = 11.84HH159 pKa = 6.15LLGTMAAGPYY169 pKa = 8.77MSMVKK174 pKa = 10.54GLDD177 pKa = 3.36KK178 pKa = 10.95RR179 pKa = 11.84DD180 pKa = 3.48GGVLLGMGPLSDD192 pKa = 3.77SYY194 pKa = 11.64QRR196 pKa = 11.84IADD199 pKa = 3.62WCKK202 pKa = 10.13GADD205 pKa = 3.31SFIFLDD211 pKa = 4.77FSKK214 pKa = 10.62FDD216 pKa = 3.29QRR218 pKa = 11.84LPRR221 pKa = 11.84DD222 pKa = 3.47LLQQVMKK229 pKa = 10.27HH230 pKa = 4.32IRR232 pKa = 11.84GRR234 pKa = 11.84FDD236 pKa = 3.18NVPGGRR242 pKa = 11.84AYY244 pKa = 9.54WEE246 pKa = 4.48SEE248 pKa = 4.02CEE250 pKa = 3.96QLIDD254 pKa = 3.82TQIAMPDD261 pKa = 3.23GNVYY265 pKa = 10.45RR266 pKa = 11.84KK267 pKa = 9.64SQGVASGDD275 pKa = 3.17PWTSIVGSYY284 pKa = 10.96ANWIILRR291 pKa = 11.84RR292 pKa = 11.84VCDD295 pKa = 3.78VLGLRR300 pKa = 11.84AKK302 pKa = 9.93IWTFGDD308 pKa = 3.42DD309 pKa = 4.26SIIAVYY315 pKa = 8.66GQKK318 pKa = 10.07VGSGIIPRR326 pKa = 11.84IAEE329 pKa = 4.0VALSEE334 pKa = 4.82FNMQVSMEE342 pKa = 4.02KK343 pKa = 10.21SYY345 pKa = 11.84ASTTLVDD352 pKa = 4.06IDD354 pKa = 5.42DD355 pKa = 4.71DD356 pKa = 4.42PEE358 pKa = 4.14PHH360 pKa = 6.37VSGSFLSLYY369 pKa = 10.3FLQTPMGVRR378 pKa = 11.84PTRR381 pKa = 11.84PLQDD385 pKa = 3.44LHH387 pKa = 8.56EE388 pKa = 4.63MFLVPEE394 pKa = 4.55RR395 pKa = 11.84NRR397 pKa = 11.84KK398 pKa = 7.88DD399 pKa = 3.56LRR401 pKa = 11.84WEE403 pKa = 3.77IVRR406 pKa = 11.84TSMAYY411 pKa = 10.12LVFYY415 pKa = 10.58YY416 pKa = 10.34NEE418 pKa = 3.76KK419 pKa = 9.75ARR421 pKa = 11.84YY422 pKa = 9.02VIEE425 pKa = 5.23EE426 pKa = 4.04YY427 pKa = 10.14WDD429 pKa = 4.02WIHH432 pKa = 6.24TRR434 pKa = 11.84YY435 pKa = 9.72KK436 pKa = 10.28VPQLRR441 pKa = 11.84GTAEE445 pKa = 4.33DD446 pKa = 3.61LTLLRR451 pKa = 11.84EE452 pKa = 4.02MDD454 pKa = 3.82IPWSAFKK461 pKa = 10.6WEE463 pKa = 3.88WLEE466 pKa = 3.69RR467 pKa = 11.84LPRR470 pKa = 11.84PGEE473 pKa = 3.89VEE475 pKa = 4.29LLYY478 pKa = 10.71KK479 pKa = 10.52YY480 pKa = 10.56GHH482 pKa = 6.58TGFFPPSLWGAWYY495 pKa = 10.48SKK497 pKa = 11.32YY498 pKa = 11.0DD499 pKa = 4.92DD500 pKa = 5.78DD501 pKa = 6.02DD502 pKa = 4.6LGNRR506 pKa = 11.84VSLPMRR512 pKa = 11.84TPQGSDD518 pKa = 2.59HH519 pKa = 6.97GGGPVNN525 pKa = 4.16
MM1 pKa = 7.29TCMAGPSEE9 pKa = 4.35EE10 pKa = 4.17VVMSEE15 pKa = 4.29FVPPAISDD23 pKa = 4.07LYY25 pKa = 11.28KK26 pKa = 10.89HH27 pKa = 6.0MEE29 pKa = 4.12GFGNRR34 pKa = 11.84NPGRR38 pKa = 11.84VKK40 pKa = 9.91PTTDD44 pKa = 2.67TYY46 pKa = 11.75AGLFSEE52 pKa = 4.69LRR54 pKa = 11.84AVEE57 pKa = 3.93GWTRR61 pKa = 11.84SISGWLDD68 pKa = 3.06PALLPKK74 pKa = 10.11IVVPSQTSPGIRR86 pKa = 11.84WKK88 pKa = 10.76RR89 pKa = 11.84LGYY92 pKa = 7.65RR93 pKa = 11.84TKK95 pKa = 10.6RR96 pKa = 11.84AALMPAVVEE105 pKa = 4.05ASRR108 pKa = 11.84LVRR111 pKa = 11.84RR112 pKa = 11.84MADD115 pKa = 2.77TGEE118 pKa = 4.23KK119 pKa = 10.75YY120 pKa = 9.59EE121 pKa = 4.06VPPAGVAGRR130 pKa = 11.84GKK132 pKa = 10.54RR133 pKa = 11.84LDD135 pKa = 3.42SCRR138 pKa = 11.84DD139 pKa = 3.1TNAPDD144 pKa = 3.4RR145 pKa = 11.84KK146 pKa = 9.02EE147 pKa = 3.67GRR149 pKa = 11.84LIVMPDD155 pKa = 3.6LVRR158 pKa = 11.84HH159 pKa = 6.15LLGTMAAGPYY169 pKa = 8.77MSMVKK174 pKa = 10.54GLDD177 pKa = 3.36KK178 pKa = 10.95RR179 pKa = 11.84DD180 pKa = 3.48GGVLLGMGPLSDD192 pKa = 3.77SYY194 pKa = 11.64QRR196 pKa = 11.84IADD199 pKa = 3.62WCKK202 pKa = 10.13GADD205 pKa = 3.31SFIFLDD211 pKa = 4.77FSKK214 pKa = 10.62FDD216 pKa = 3.29QRR218 pKa = 11.84LPRR221 pKa = 11.84DD222 pKa = 3.47LLQQVMKK229 pKa = 10.27HH230 pKa = 4.32IRR232 pKa = 11.84GRR234 pKa = 11.84FDD236 pKa = 3.18NVPGGRR242 pKa = 11.84AYY244 pKa = 9.54WEE246 pKa = 4.48SEE248 pKa = 4.02CEE250 pKa = 3.96QLIDD254 pKa = 3.82TQIAMPDD261 pKa = 3.23GNVYY265 pKa = 10.45RR266 pKa = 11.84KK267 pKa = 9.64SQGVASGDD275 pKa = 3.17PWTSIVGSYY284 pKa = 10.96ANWIILRR291 pKa = 11.84RR292 pKa = 11.84VCDD295 pKa = 3.78VLGLRR300 pKa = 11.84AKK302 pKa = 9.93IWTFGDD308 pKa = 3.42DD309 pKa = 4.26SIIAVYY315 pKa = 8.66GQKK318 pKa = 10.07VGSGIIPRR326 pKa = 11.84IAEE329 pKa = 4.0VALSEE334 pKa = 4.82FNMQVSMEE342 pKa = 4.02KK343 pKa = 10.21SYY345 pKa = 11.84ASTTLVDD352 pKa = 4.06IDD354 pKa = 5.42DD355 pKa = 4.71DD356 pKa = 4.42PEE358 pKa = 4.14PHH360 pKa = 6.37VSGSFLSLYY369 pKa = 10.3FLQTPMGVRR378 pKa = 11.84PTRR381 pKa = 11.84PLQDD385 pKa = 3.44LHH387 pKa = 8.56EE388 pKa = 4.63MFLVPEE394 pKa = 4.55RR395 pKa = 11.84NRR397 pKa = 11.84KK398 pKa = 7.88DD399 pKa = 3.56LRR401 pKa = 11.84WEE403 pKa = 3.77IVRR406 pKa = 11.84TSMAYY411 pKa = 10.12LVFYY415 pKa = 10.58YY416 pKa = 10.34NEE418 pKa = 3.76KK419 pKa = 9.75ARR421 pKa = 11.84YY422 pKa = 9.02VIEE425 pKa = 5.23EE426 pKa = 4.04YY427 pKa = 10.14WDD429 pKa = 4.02WIHH432 pKa = 6.24TRR434 pKa = 11.84YY435 pKa = 9.72KK436 pKa = 10.28VPQLRR441 pKa = 11.84GTAEE445 pKa = 4.33DD446 pKa = 3.61LTLLRR451 pKa = 11.84EE452 pKa = 4.02MDD454 pKa = 3.82IPWSAFKK461 pKa = 10.6WEE463 pKa = 3.88WLEE466 pKa = 3.69RR467 pKa = 11.84LPRR470 pKa = 11.84PGEE473 pKa = 3.89VEE475 pKa = 4.29LLYY478 pKa = 10.71KK479 pKa = 10.52YY480 pKa = 10.56GHH482 pKa = 6.58TGFFPPSLWGAWYY495 pKa = 10.48SKK497 pKa = 11.32YY498 pKa = 11.0DD499 pKa = 4.92DD500 pKa = 5.78DD501 pKa = 6.02DD502 pKa = 4.6LGNRR506 pKa = 11.84VSLPMRR512 pKa = 11.84TPQGSDD518 pKa = 2.59HH519 pKa = 6.97GGGPVNN525 pKa = 4.16
Molecular weight: 59.54 kDa
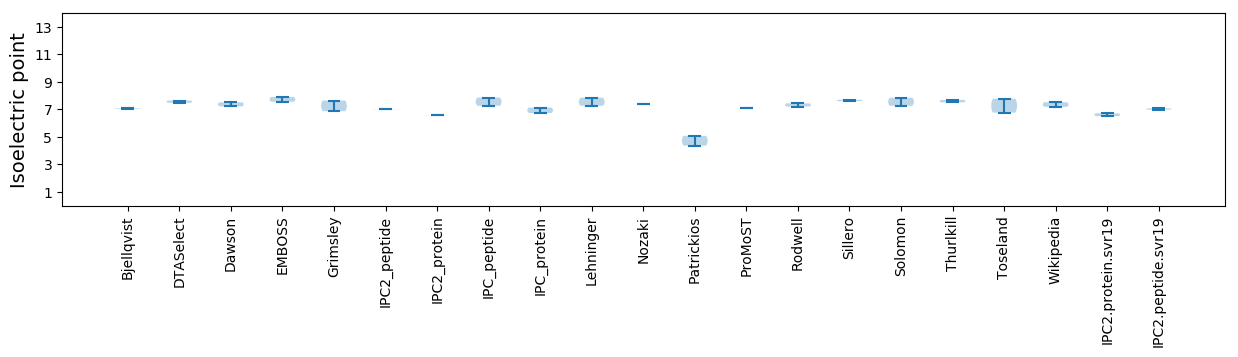
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3ZCM4|A0A0G3ZCM4_9VIRU Uncharacterized protein OS=Ustilaginoidea virens unassigned RNA virus HNND-1 OX=1670975 PE=4 SV=1
MM1 pKa = 7.62SSTHH5 pKa = 5.25TPKK8 pKa = 8.43TTAEE12 pKa = 4.12VKK14 pKa = 9.86PVSSSLPAVGTPAQKK29 pKa = 10.78GAGAKK34 pKa = 9.4GKK36 pKa = 9.78RR37 pKa = 11.84VSGSSTASGGVRR49 pKa = 11.84EE50 pKa = 4.99GFDD53 pKa = 4.25PDD55 pKa = 3.9PEE57 pKa = 4.5SALSVNWASGYY68 pKa = 10.31GVSTPAVTLDD78 pKa = 3.52IIRR81 pKa = 11.84RR82 pKa = 11.84FLMHH86 pKa = 6.32QGVSALPRR94 pKa = 11.84GVEE97 pKa = 4.38DD98 pKa = 5.28LFPSDD103 pKa = 2.77WRR105 pKa = 11.84VGKK108 pKa = 8.52TPVPLEE114 pKa = 3.69WDD116 pKa = 3.93TVGGRR121 pKa = 11.84ALAGVLPNIRR131 pKa = 11.84AYY133 pKa = 10.5DD134 pKa = 3.39VDD136 pKa = 3.14GRR138 pKa = 11.84EE139 pKa = 3.99IHH141 pKa = 5.91WVGEE145 pKa = 4.12GRR147 pKa = 11.84DD148 pKa = 3.36ISHH151 pKa = 7.77ASRR154 pKa = 11.84LKK156 pKa = 10.97AEE158 pKa = 3.86WDD160 pKa = 3.47APITIVAAILTAVKK174 pKa = 9.84KK175 pKa = 10.84AKK177 pKa = 10.11ISEE180 pKa = 4.14EE181 pKa = 4.01DD182 pKa = 3.42VKK184 pKa = 11.19LVAQHH189 pKa = 6.53SPQALEE195 pKa = 3.88KK196 pKa = 10.66LKK198 pKa = 10.78EE199 pKa = 3.8AGAAYY204 pKa = 9.09KK205 pKa = 10.68AHH207 pKa = 7.29LNRR210 pKa = 11.84FLDD213 pKa = 4.18PVALRR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.51PRR222 pKa = 11.84YY223 pKa = 10.18GEE225 pKa = 3.59VAARR229 pKa = 11.84YY230 pKa = 8.51KK231 pKa = 11.07AEE233 pKa = 3.76IRR235 pKa = 11.84SLVAEE240 pKa = 4.12QQAKK244 pKa = 9.33IVEE247 pKa = 4.18ARR249 pKa = 11.84KK250 pKa = 8.12VTRR253 pKa = 11.84RR254 pKa = 11.84LNQLLGQRR262 pKa = 11.84DD263 pKa = 3.74EE264 pKa = 4.77EE265 pKa = 4.41LAKK268 pKa = 10.35LDD270 pKa = 4.09PGFTPKK276 pKa = 10.45RR277 pKa = 11.84ATAAAEE283 pKa = 3.83LAKK286 pKa = 10.72FGITLEE292 pKa = 4.21EE293 pKa = 4.51GEE295 pKa = 4.32EE296 pKa = 4.21AEE298 pKa = 4.31SAEE301 pKa = 4.44AGLTAQMLEE310 pKa = 4.33DD311 pKa = 4.29LNFF314 pKa = 3.84
MM1 pKa = 7.62SSTHH5 pKa = 5.25TPKK8 pKa = 8.43TTAEE12 pKa = 4.12VKK14 pKa = 9.86PVSSSLPAVGTPAQKK29 pKa = 10.78GAGAKK34 pKa = 9.4GKK36 pKa = 9.78RR37 pKa = 11.84VSGSSTASGGVRR49 pKa = 11.84EE50 pKa = 4.99GFDD53 pKa = 4.25PDD55 pKa = 3.9PEE57 pKa = 4.5SALSVNWASGYY68 pKa = 10.31GVSTPAVTLDD78 pKa = 3.52IIRR81 pKa = 11.84RR82 pKa = 11.84FLMHH86 pKa = 6.32QGVSALPRR94 pKa = 11.84GVEE97 pKa = 4.38DD98 pKa = 5.28LFPSDD103 pKa = 2.77WRR105 pKa = 11.84VGKK108 pKa = 8.52TPVPLEE114 pKa = 3.69WDD116 pKa = 3.93TVGGRR121 pKa = 11.84ALAGVLPNIRR131 pKa = 11.84AYY133 pKa = 10.5DD134 pKa = 3.39VDD136 pKa = 3.14GRR138 pKa = 11.84EE139 pKa = 3.99IHH141 pKa = 5.91WVGEE145 pKa = 4.12GRR147 pKa = 11.84DD148 pKa = 3.36ISHH151 pKa = 7.77ASRR154 pKa = 11.84LKK156 pKa = 10.97AEE158 pKa = 3.86WDD160 pKa = 3.47APITIVAAILTAVKK174 pKa = 9.84KK175 pKa = 10.84AKK177 pKa = 10.11ISEE180 pKa = 4.14EE181 pKa = 4.01DD182 pKa = 3.42VKK184 pKa = 11.19LVAQHH189 pKa = 6.53SPQALEE195 pKa = 3.88KK196 pKa = 10.66LKK198 pKa = 10.78EE199 pKa = 3.8AGAAYY204 pKa = 9.09KK205 pKa = 10.68AHH207 pKa = 7.29LNRR210 pKa = 11.84FLDD213 pKa = 4.18PVALRR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.51PRR222 pKa = 11.84YY223 pKa = 10.18GEE225 pKa = 3.59VAARR229 pKa = 11.84YY230 pKa = 8.51KK231 pKa = 11.07AEE233 pKa = 3.76IRR235 pKa = 11.84SLVAEE240 pKa = 4.12QQAKK244 pKa = 9.33IVEE247 pKa = 4.18ARR249 pKa = 11.84KK250 pKa = 8.12VTRR253 pKa = 11.84RR254 pKa = 11.84LNQLLGQRR262 pKa = 11.84DD263 pKa = 3.74EE264 pKa = 4.77EE265 pKa = 4.41LAKK268 pKa = 10.35LDD270 pKa = 4.09PGFTPKK276 pKa = 10.45RR277 pKa = 11.84ATAAAEE283 pKa = 3.83LAKK286 pKa = 10.72FGITLEE292 pKa = 4.21EE293 pKa = 4.51GEE295 pKa = 4.32EE296 pKa = 4.21AEE298 pKa = 4.31SAEE301 pKa = 4.44AGLTAQMLEE310 pKa = 4.33DD311 pKa = 4.29LNFF314 pKa = 3.84
Molecular weight: 33.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
839 |
314 |
525 |
419.5 |
46.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.82 ± 2.828 | 0.596 ± 0.346 |
6.317 ± 0.709 | 6.555 ± 1.001 |
2.861 ± 0.366 | 8.582 ± 0.175 |
1.669 ± 0.141 | 4.291 ± 0.272 |
5.125 ± 0.722 | 8.939 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.741 ± 1.036 | 1.907 ± 0.183 |
6.436 ± 0.408 | 2.741 ± 0.072 |
7.747 ± 0.43 | 6.675 ± 0.008 |
4.768 ± 0.375 | 7.509 ± 0.263 |
2.503 ± 0.528 | 3.218 ± 0.943 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
