
Ustilaginoidea virens nonsegmented virus 2
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
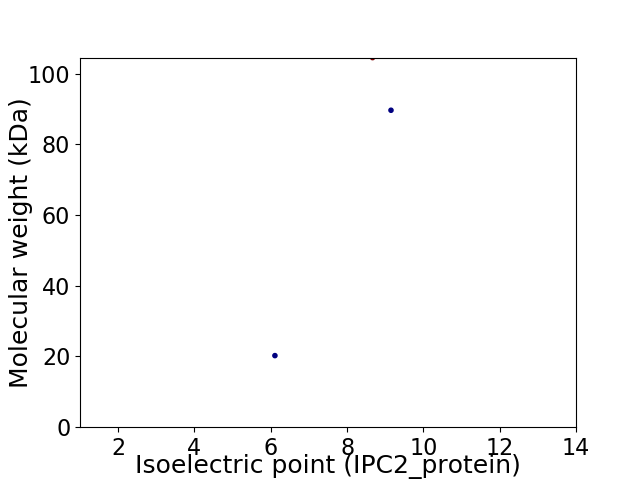
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
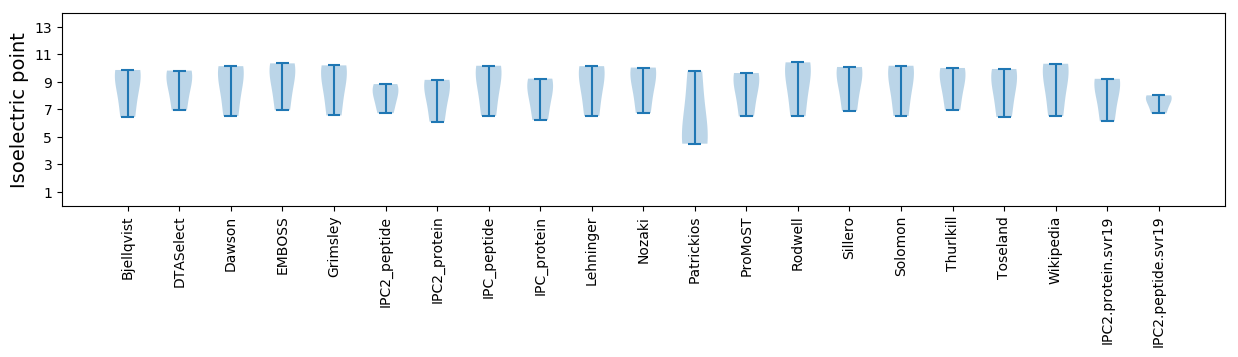
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346P9B8|A0A346P9B8_9VIRU RNA-dependent RNA polymerase OS=Ustilaginoidea virens nonsegmented virus 2 OX=2305465 PE=4 SV=1
MM1 pKa = 7.81HH2 pKa = 7.0YY3 pKa = 9.98HH4 pKa = 6.67LCSPPHH10 pKa = 6.72AMADD14 pKa = 3.17KK15 pKa = 10.83ALQQEE20 pKa = 4.74LEE22 pKa = 4.17LEE24 pKa = 4.3ALVKK28 pKa = 10.75AFNEE32 pKa = 4.12NFKK35 pKa = 8.63TTAGPDD41 pKa = 3.39AVKK44 pKa = 10.64KK45 pKa = 10.76LLGSPHH51 pKa = 4.78VRR53 pKa = 11.84GWYY56 pKa = 9.82AEE58 pKa = 3.85YY59 pKa = 10.27RR60 pKa = 11.84QEE62 pKa = 3.94QAKK65 pKa = 10.45EE66 pKa = 3.75KK67 pKa = 10.63GLPEE71 pKa = 4.04DD72 pKa = 3.34WAAVGLTAQEE82 pKa = 3.69WAEE85 pKa = 4.0AQSARR90 pKa = 11.84SLRR93 pKa = 11.84RR94 pKa = 11.84EE95 pKa = 3.95ARR97 pKa = 11.84AEE99 pKa = 3.87AAKK102 pKa = 9.65TLAAEE107 pKa = 4.49AARR110 pKa = 11.84YY111 pKa = 8.2DD112 pKa = 3.78AAVKK116 pKa = 10.36RR117 pKa = 11.84INATCAAALDD127 pKa = 4.04RR128 pKa = 11.84AAAPMVLALEE138 pKa = 4.65GGYY141 pKa = 9.64EE142 pKa = 4.02QLPLAAQIRR151 pKa = 11.84VEE153 pKa = 4.24SSADD157 pKa = 3.11PEE159 pKa = 4.36ARR161 pKa = 11.84AKK163 pKa = 10.24CLKK166 pKa = 10.8AEE168 pKa = 4.22LADD171 pKa = 4.56ARR173 pKa = 11.84AVWVAKK179 pKa = 10.34ARR181 pKa = 11.84AGEE184 pKa = 4.08APFF187 pKa = 4.75
MM1 pKa = 7.81HH2 pKa = 7.0YY3 pKa = 9.98HH4 pKa = 6.67LCSPPHH10 pKa = 6.72AMADD14 pKa = 3.17KK15 pKa = 10.83ALQQEE20 pKa = 4.74LEE22 pKa = 4.17LEE24 pKa = 4.3ALVKK28 pKa = 10.75AFNEE32 pKa = 4.12NFKK35 pKa = 8.63TTAGPDD41 pKa = 3.39AVKK44 pKa = 10.64KK45 pKa = 10.76LLGSPHH51 pKa = 4.78VRR53 pKa = 11.84GWYY56 pKa = 9.82AEE58 pKa = 3.85YY59 pKa = 10.27RR60 pKa = 11.84QEE62 pKa = 3.94QAKK65 pKa = 10.45EE66 pKa = 3.75KK67 pKa = 10.63GLPEE71 pKa = 4.04DD72 pKa = 3.34WAAVGLTAQEE82 pKa = 3.69WAEE85 pKa = 4.0AQSARR90 pKa = 11.84SLRR93 pKa = 11.84RR94 pKa = 11.84EE95 pKa = 3.95ARR97 pKa = 11.84AEE99 pKa = 3.87AAKK102 pKa = 9.65TLAAEE107 pKa = 4.49AARR110 pKa = 11.84YY111 pKa = 8.2DD112 pKa = 3.78AAVKK116 pKa = 10.36RR117 pKa = 11.84INATCAAALDD127 pKa = 4.04RR128 pKa = 11.84AAAPMVLALEE138 pKa = 4.65GGYY141 pKa = 9.64EE142 pKa = 4.02QLPLAAQIRR151 pKa = 11.84VEE153 pKa = 4.24SSADD157 pKa = 3.11PEE159 pKa = 4.36ARR161 pKa = 11.84AKK163 pKa = 10.24CLKK166 pKa = 10.8AEE168 pKa = 4.22LADD171 pKa = 4.56ARR173 pKa = 11.84AVWVAKK179 pKa = 10.34ARR181 pKa = 11.84AGEE184 pKa = 4.08APFF187 pKa = 4.75
Molecular weight: 20.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346P9B9|A0A346P9B9_9VIRU Fusion protein OS=Ustilaginoidea virens nonsegmented virus 2 OX=2305465 PE=4 SV=1
MM1 pKa = 8.03SSFRR5 pKa = 11.84WLLRR9 pKa = 11.84SGWNPQRR16 pKa = 11.84TPRR19 pKa = 11.84HH20 pKa = 5.32GRR22 pKa = 11.84SVSKK26 pKa = 10.27PSSRR30 pKa = 11.84THH32 pKa = 5.77VPCGLLKK39 pKa = 10.63PGLARR44 pKa = 11.84LPFSLPMPLDD54 pKa = 4.12GPTWSCPAYY63 pKa = 10.33GLLDD67 pKa = 4.35RR68 pKa = 11.84GTLTAVVSRR77 pKa = 11.84LSRR80 pKa = 11.84KK81 pKa = 5.14WHH83 pKa = 6.34RR84 pKa = 11.84LRR86 pKa = 11.84EE87 pKa = 4.5SVFSRR92 pKa = 11.84VPCDD96 pKa = 3.3SPQSHH101 pKa = 6.36VKK103 pKa = 10.22GQLHH107 pKa = 5.8MFWSARR113 pKa = 11.84LDD115 pKa = 3.42EE116 pKa = 4.62AVEE119 pKa = 3.87HH120 pKa = 6.74RR121 pKa = 11.84EE122 pKa = 4.07TYY124 pKa = 10.86KK125 pKa = 10.79LVQSLPEE132 pKa = 3.69TDD134 pKa = 4.42RR135 pKa = 11.84IRR137 pKa = 11.84MAWKK141 pKa = 9.17YY142 pKa = 10.63LNRR145 pKa = 11.84RR146 pKa = 11.84KK147 pKa = 10.03AQAAKK152 pKa = 10.65ARR154 pKa = 11.84DD155 pKa = 4.0DD156 pKa = 3.68SSGSHH161 pKa = 6.9LPRR164 pKa = 11.84VQAAALWLVLDD175 pKa = 5.03DD176 pKa = 6.05FSLTPARR183 pKa = 11.84ATVLAHH189 pKa = 6.34AEE191 pKa = 3.88YY192 pKa = 10.12RR193 pKa = 11.84RR194 pKa = 11.84KK195 pKa = 10.18RR196 pKa = 11.84FEE198 pKa = 3.71KK199 pKa = 9.99PEE201 pKa = 3.85RR202 pKa = 11.84KK203 pKa = 9.22IRR205 pKa = 11.84TTEE208 pKa = 3.85WVGGAPNPIALTSEE222 pKa = 4.27RR223 pKa = 11.84QVSTKK228 pKa = 7.92WKK230 pKa = 8.81RR231 pKa = 11.84AQSNAEE237 pKa = 3.99RR238 pKa = 11.84LAALCDD244 pKa = 3.98DD245 pKa = 4.63APFPPTAVARR255 pKa = 11.84LDD257 pKa = 3.55GRR259 pKa = 11.84LEE261 pKa = 4.18LPSGCRR267 pKa = 11.84PQKK270 pKa = 10.27FVLPGGPALEE280 pKa = 5.33FIRR283 pKa = 11.84EE284 pKa = 4.01RR285 pKa = 11.84RR286 pKa = 11.84PSLLKK291 pKa = 10.23WLGPSDD297 pKa = 3.78ACVGGSWDD305 pKa = 3.36TSKK308 pKa = 10.7WSFEE312 pKa = 4.39VACQPNKK319 pKa = 10.64GSGHH323 pKa = 6.66AGDD326 pKa = 5.19LYY328 pKa = 10.74RR329 pKa = 11.84ALSSGYY335 pKa = 10.19KK336 pKa = 9.68DD337 pKa = 3.7LKK339 pKa = 10.5LGRR342 pKa = 11.84LPQPSIRR349 pKa = 11.84ALEE352 pKa = 4.1CVKK355 pKa = 10.8VNGRR359 pKa = 11.84AYY361 pKa = 9.83PGIFSSRR368 pKa = 11.84IGNNRR373 pKa = 11.84KK374 pKa = 6.57TAYY377 pKa = 8.98GACVEE382 pKa = 4.37IAKK385 pKa = 9.16QHH387 pKa = 5.67YY388 pKa = 9.57LDD390 pKa = 3.47ARR392 pKa = 11.84EE393 pKa = 4.01RR394 pKa = 11.84FEE396 pKa = 5.48PDD398 pKa = 2.76LSLWACGGRR407 pKa = 11.84GKK409 pKa = 9.64PSQMVQVGDD418 pKa = 3.73RR419 pKa = 11.84LKK421 pKa = 11.06SRR423 pKa = 11.84LILMPEE429 pKa = 4.16TPSALLEE436 pKa = 4.27SAFAQPFTAMLSAVRR451 pKa = 11.84GDD453 pKa = 3.51IMIGASMTDD462 pKa = 2.59RR463 pKa = 11.84GFRR466 pKa = 11.84RR467 pKa = 11.84VLSPVEE473 pKa = 4.22DD474 pKa = 3.46AHH476 pKa = 6.83HH477 pKa = 6.41VKK479 pKa = 10.86AFDD482 pKa = 3.09WSGFDD487 pKa = 3.34SRR489 pKa = 11.84VRR491 pKa = 11.84EE492 pKa = 4.02DD493 pKa = 3.62MIVTAFGIVRR503 pKa = 11.84ACFKK507 pKa = 11.16GDD509 pKa = 4.18DD510 pKa = 3.5AWLDD514 pKa = 3.57NVFLRR519 pKa = 11.84FISHH523 pKa = 6.8FLVKK527 pKa = 10.24RR528 pKa = 11.84VVTPGGWLYY537 pKa = 10.31TLANGVPSGSPFTSIIDD554 pKa = 3.73SLVNWLVITDD564 pKa = 4.85LEE566 pKa = 4.23ICMGGLSAPTKK577 pKa = 9.67NRR579 pKa = 11.84RR580 pKa = 11.84RR581 pKa = 11.84VYY583 pKa = 11.07GDD585 pKa = 4.82DD586 pKa = 3.24FMQAFFAPCLEE597 pKa = 3.94RR598 pKa = 11.84DD599 pKa = 3.62AYY601 pKa = 9.8IALAFEE607 pKa = 4.16RR608 pKa = 11.84WGFVAKK614 pKa = 10.25PSAALEE620 pKa = 4.33GVACANTADD629 pKa = 3.72ASLPFLSFRR638 pKa = 11.84FPFGLPARR646 pKa = 11.84PIQDD650 pKa = 3.28ALKK653 pKa = 10.5IGLLPQKK660 pKa = 10.61ARR662 pKa = 11.84YY663 pKa = 9.16SYY665 pKa = 9.68SAQAARR671 pKa = 11.84VCYY674 pKa = 9.85LDD676 pKa = 4.94HH677 pKa = 6.89FAPYY681 pKa = 10.34DD682 pKa = 3.65PEE684 pKa = 4.26TIEE687 pKa = 3.77YY688 pKa = 9.22HH689 pKa = 6.39RR690 pKa = 11.84EE691 pKa = 3.75YY692 pKa = 11.01FNWLQTKK699 pKa = 9.86IPGMTWADD707 pKa = 3.52GRR709 pKa = 11.84PQPDD713 pKa = 3.87LVSPWIHH720 pKa = 6.0KK721 pKa = 10.85AMVNFVAAGFAPGVVSLGEE740 pKa = 4.0WFRR743 pKa = 11.84QEE745 pKa = 5.18DD746 pKa = 3.67PRR748 pKa = 11.84RR749 pKa = 11.84WPDD752 pKa = 2.29RR753 pKa = 11.84WVPRR757 pKa = 11.84RR758 pKa = 11.84CGRR761 pKa = 11.84LVPRR765 pKa = 11.84AAWSQGKK772 pKa = 8.49LQSALSTLRR781 pKa = 11.84WGNCAEE787 pKa = 4.21STFARR792 pKa = 11.84LRR794 pKa = 11.84WKK796 pKa = 9.11TLL798 pKa = 3.07
MM1 pKa = 8.03SSFRR5 pKa = 11.84WLLRR9 pKa = 11.84SGWNPQRR16 pKa = 11.84TPRR19 pKa = 11.84HH20 pKa = 5.32GRR22 pKa = 11.84SVSKK26 pKa = 10.27PSSRR30 pKa = 11.84THH32 pKa = 5.77VPCGLLKK39 pKa = 10.63PGLARR44 pKa = 11.84LPFSLPMPLDD54 pKa = 4.12GPTWSCPAYY63 pKa = 10.33GLLDD67 pKa = 4.35RR68 pKa = 11.84GTLTAVVSRR77 pKa = 11.84LSRR80 pKa = 11.84KK81 pKa = 5.14WHH83 pKa = 6.34RR84 pKa = 11.84LRR86 pKa = 11.84EE87 pKa = 4.5SVFSRR92 pKa = 11.84VPCDD96 pKa = 3.3SPQSHH101 pKa = 6.36VKK103 pKa = 10.22GQLHH107 pKa = 5.8MFWSARR113 pKa = 11.84LDD115 pKa = 3.42EE116 pKa = 4.62AVEE119 pKa = 3.87HH120 pKa = 6.74RR121 pKa = 11.84EE122 pKa = 4.07TYY124 pKa = 10.86KK125 pKa = 10.79LVQSLPEE132 pKa = 3.69TDD134 pKa = 4.42RR135 pKa = 11.84IRR137 pKa = 11.84MAWKK141 pKa = 9.17YY142 pKa = 10.63LNRR145 pKa = 11.84RR146 pKa = 11.84KK147 pKa = 10.03AQAAKK152 pKa = 10.65ARR154 pKa = 11.84DD155 pKa = 4.0DD156 pKa = 3.68SSGSHH161 pKa = 6.9LPRR164 pKa = 11.84VQAAALWLVLDD175 pKa = 5.03DD176 pKa = 6.05FSLTPARR183 pKa = 11.84ATVLAHH189 pKa = 6.34AEE191 pKa = 3.88YY192 pKa = 10.12RR193 pKa = 11.84RR194 pKa = 11.84KK195 pKa = 10.18RR196 pKa = 11.84FEE198 pKa = 3.71KK199 pKa = 9.99PEE201 pKa = 3.85RR202 pKa = 11.84KK203 pKa = 9.22IRR205 pKa = 11.84TTEE208 pKa = 3.85WVGGAPNPIALTSEE222 pKa = 4.27RR223 pKa = 11.84QVSTKK228 pKa = 7.92WKK230 pKa = 8.81RR231 pKa = 11.84AQSNAEE237 pKa = 3.99RR238 pKa = 11.84LAALCDD244 pKa = 3.98DD245 pKa = 4.63APFPPTAVARR255 pKa = 11.84LDD257 pKa = 3.55GRR259 pKa = 11.84LEE261 pKa = 4.18LPSGCRR267 pKa = 11.84PQKK270 pKa = 10.27FVLPGGPALEE280 pKa = 5.33FIRR283 pKa = 11.84EE284 pKa = 4.01RR285 pKa = 11.84RR286 pKa = 11.84PSLLKK291 pKa = 10.23WLGPSDD297 pKa = 3.78ACVGGSWDD305 pKa = 3.36TSKK308 pKa = 10.7WSFEE312 pKa = 4.39VACQPNKK319 pKa = 10.64GSGHH323 pKa = 6.66AGDD326 pKa = 5.19LYY328 pKa = 10.74RR329 pKa = 11.84ALSSGYY335 pKa = 10.19KK336 pKa = 9.68DD337 pKa = 3.7LKK339 pKa = 10.5LGRR342 pKa = 11.84LPQPSIRR349 pKa = 11.84ALEE352 pKa = 4.1CVKK355 pKa = 10.8VNGRR359 pKa = 11.84AYY361 pKa = 9.83PGIFSSRR368 pKa = 11.84IGNNRR373 pKa = 11.84KK374 pKa = 6.57TAYY377 pKa = 8.98GACVEE382 pKa = 4.37IAKK385 pKa = 9.16QHH387 pKa = 5.67YY388 pKa = 9.57LDD390 pKa = 3.47ARR392 pKa = 11.84EE393 pKa = 4.01RR394 pKa = 11.84FEE396 pKa = 5.48PDD398 pKa = 2.76LSLWACGGRR407 pKa = 11.84GKK409 pKa = 9.64PSQMVQVGDD418 pKa = 3.73RR419 pKa = 11.84LKK421 pKa = 11.06SRR423 pKa = 11.84LILMPEE429 pKa = 4.16TPSALLEE436 pKa = 4.27SAFAQPFTAMLSAVRR451 pKa = 11.84GDD453 pKa = 3.51IMIGASMTDD462 pKa = 2.59RR463 pKa = 11.84GFRR466 pKa = 11.84RR467 pKa = 11.84VLSPVEE473 pKa = 4.22DD474 pKa = 3.46AHH476 pKa = 6.83HH477 pKa = 6.41VKK479 pKa = 10.86AFDD482 pKa = 3.09WSGFDD487 pKa = 3.34SRR489 pKa = 11.84VRR491 pKa = 11.84EE492 pKa = 4.02DD493 pKa = 3.62MIVTAFGIVRR503 pKa = 11.84ACFKK507 pKa = 11.16GDD509 pKa = 4.18DD510 pKa = 3.5AWLDD514 pKa = 3.57NVFLRR519 pKa = 11.84FISHH523 pKa = 6.8FLVKK527 pKa = 10.24RR528 pKa = 11.84VVTPGGWLYY537 pKa = 10.31TLANGVPSGSPFTSIIDD554 pKa = 3.73SLVNWLVITDD564 pKa = 4.85LEE566 pKa = 4.23ICMGGLSAPTKK577 pKa = 9.67NRR579 pKa = 11.84RR580 pKa = 11.84RR581 pKa = 11.84VYY583 pKa = 11.07GDD585 pKa = 4.82DD586 pKa = 3.24FMQAFFAPCLEE597 pKa = 3.94RR598 pKa = 11.84DD599 pKa = 3.62AYY601 pKa = 9.8IALAFEE607 pKa = 4.16RR608 pKa = 11.84WGFVAKK614 pKa = 10.25PSAALEE620 pKa = 4.33GVACANTADD629 pKa = 3.72ASLPFLSFRR638 pKa = 11.84FPFGLPARR646 pKa = 11.84PIQDD650 pKa = 3.28ALKK653 pKa = 10.5IGLLPQKK660 pKa = 10.61ARR662 pKa = 11.84YY663 pKa = 9.16SYY665 pKa = 9.68SAQAARR671 pKa = 11.84VCYY674 pKa = 9.85LDD676 pKa = 4.94HH677 pKa = 6.89FAPYY681 pKa = 10.34DD682 pKa = 3.65PEE684 pKa = 4.26TIEE687 pKa = 3.77YY688 pKa = 9.22HH689 pKa = 6.39RR690 pKa = 11.84EE691 pKa = 3.75YY692 pKa = 11.01FNWLQTKK699 pKa = 9.86IPGMTWADD707 pKa = 3.52GRR709 pKa = 11.84PQPDD713 pKa = 3.87LVSPWIHH720 pKa = 6.0KK721 pKa = 10.85AMVNFVAAGFAPGVVSLGEE740 pKa = 4.0WFRR743 pKa = 11.84QEE745 pKa = 5.18DD746 pKa = 3.67PRR748 pKa = 11.84RR749 pKa = 11.84WPDD752 pKa = 2.29RR753 pKa = 11.84WVPRR757 pKa = 11.84RR758 pKa = 11.84CGRR761 pKa = 11.84LVPRR765 pKa = 11.84AAWSQGKK772 pKa = 8.49LQSALSTLRR781 pKa = 11.84WGNCAEE787 pKa = 4.21STFARR792 pKa = 11.84LRR794 pKa = 11.84WKK796 pKa = 9.11TLL798 pKa = 3.07
Molecular weight: 89.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1923 |
187 |
938 |
641.0 |
71.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.053 ± 2.626 | 2.028 ± 0.094 |
4.992 ± 0.26 | 5.408 ± 1.087 |
4.16 ± 0.566 | 6.448 ± 0.494 |
1.976 ± 0.043 | 2.704 ± 0.349 |
4.784 ± 0.365 | 9.776 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.716 ± 0.028 | 1.924 ± 0.072 |
6.76 ± 0.476 | 3.276 ± 0.229 |
9.1 ± 0.518 | 6.812 ± 0.855 |
3.744 ± 0.24 | 5.824 ± 0.216 |
3.12 ± 0.225 | 2.392 ± 0.086 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
