
Rhodococcus sp. SMB37
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus; unclassified Rhodococcus
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
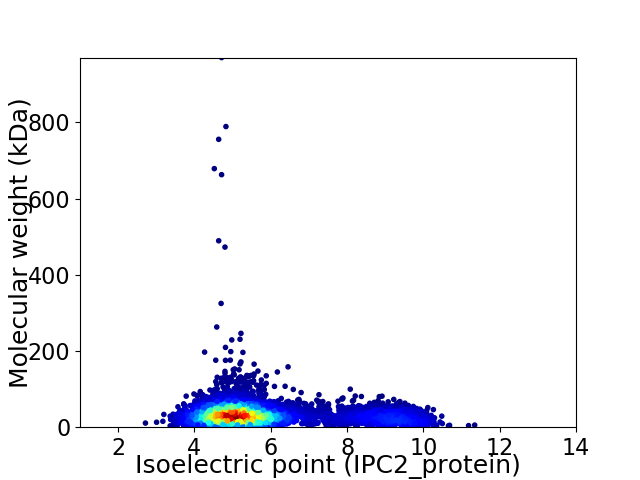
Virtual 2D-PAGE plot for 5840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R2D2J7|A0A4R2D2J7_9NOCA NADPH-dependent sulfite reductase flavoprotein alpha-component OS=Rhodococcus sp. SMB37 OX=2512213 GN=EV641_1182 PE=3 SV=1
MM1 pKa = 7.66ALNKK5 pKa = 10.07KK6 pKa = 9.88AMRR9 pKa = 11.84RR10 pKa = 11.84AVIAASSAALLVLTACGSDD29 pKa = 4.14DD30 pKa = 3.96GGSDD34 pKa = 3.22TGSDD38 pKa = 3.12TGAAASDD45 pKa = 3.97EE46 pKa = 4.1GRR48 pKa = 11.84GPITIVEE55 pKa = 4.33GQDD58 pKa = 3.43PLNAALEE65 pKa = 4.28NIIADD70 pKa = 3.96WNAEE74 pKa = 4.08NPDD77 pKa = 3.49EE78 pKa = 4.35QVTFKK83 pKa = 10.92PLARR87 pKa = 11.84EE88 pKa = 4.33ANDD91 pKa = 3.24QYY93 pKa = 12.07ADD95 pKa = 2.99IAQRR99 pKa = 11.84MQAQSDD105 pKa = 4.27EE106 pKa = 4.39YY107 pKa = 11.43DD108 pKa = 3.61LVAVDD113 pKa = 3.64VTNTAEE119 pKa = 4.07FAANGWLLPLEE130 pKa = 4.51GDD132 pKa = 3.83NAIDD136 pKa = 3.9TDD138 pKa = 4.16GLLPATVEE146 pKa = 3.99SGTYY150 pKa = 10.44NGTLYY155 pKa = 10.8AAPKK159 pKa = 7.51NTNGAFLYY167 pKa = 10.7YY168 pKa = 9.71RR169 pKa = 11.84TDD171 pKa = 3.75LVDD174 pKa = 3.45QAPATWDD181 pKa = 3.43EE182 pKa = 4.75LKK184 pKa = 11.01AEE186 pKa = 4.28CPKK189 pKa = 10.96AEE191 pKa = 4.19EE192 pKa = 4.57AGIACYY198 pKa = 9.96IGQFSNYY205 pKa = 9.48EE206 pKa = 3.97GLTVNVTEE214 pKa = 4.55IVNAFGGSFVAEE226 pKa = 4.84DD227 pKa = 4.29GLTPAIDD234 pKa = 3.54EE235 pKa = 4.26QTAEE239 pKa = 4.14GLQFIVDD246 pKa = 3.93SFEE249 pKa = 5.64DD250 pKa = 3.87GTIPAAAQTYY260 pKa = 8.75TEE262 pKa = 5.06GEE264 pKa = 4.25SQTAFGNGEE273 pKa = 4.26ALFMRR278 pKa = 11.84TWPGFYY284 pKa = 9.75TDD286 pKa = 4.35GEE288 pKa = 4.45EE289 pKa = 4.0TSVAGNYY296 pKa = 9.63GIAVLPGVDD305 pKa = 3.68GPGVSTLGGYY315 pKa = 9.4NVGISAFTDD324 pKa = 3.66APATARR330 pKa = 11.84DD331 pKa = 3.91FLEE334 pKa = 4.71FMQTRR339 pKa = 11.84QSQEE343 pKa = 3.6YY344 pKa = 7.74YY345 pKa = 10.87AEE347 pKa = 4.22VGGAPVLAEE356 pKa = 4.27VYY358 pKa = 10.24DD359 pKa = 4.1DD360 pKa = 4.79PEE362 pKa = 4.43ILEE365 pKa = 4.08QYY367 pKa = 10.45PYY369 pKa = 11.08FSTLKK374 pKa = 10.32EE375 pKa = 3.98ALEE378 pKa = 4.35SAKK381 pKa = 10.02PRR383 pKa = 11.84PSSPYY388 pKa = 8.62YY389 pKa = 10.1SAVSKK394 pKa = 10.81AISDD398 pKa = 3.6NAYY401 pKa = 10.14AAIRR405 pKa = 11.84GEE407 pKa = 4.21KK408 pKa = 9.65PVQQALDD415 pKa = 3.62DD416 pKa = 4.03TQAGIEE422 pKa = 4.18TAGNN426 pKa = 3.39
MM1 pKa = 7.66ALNKK5 pKa = 10.07KK6 pKa = 9.88AMRR9 pKa = 11.84RR10 pKa = 11.84AVIAASSAALLVLTACGSDD29 pKa = 4.14DD30 pKa = 3.96GGSDD34 pKa = 3.22TGSDD38 pKa = 3.12TGAAASDD45 pKa = 3.97EE46 pKa = 4.1GRR48 pKa = 11.84GPITIVEE55 pKa = 4.33GQDD58 pKa = 3.43PLNAALEE65 pKa = 4.28NIIADD70 pKa = 3.96WNAEE74 pKa = 4.08NPDD77 pKa = 3.49EE78 pKa = 4.35QVTFKK83 pKa = 10.92PLARR87 pKa = 11.84EE88 pKa = 4.33ANDD91 pKa = 3.24QYY93 pKa = 12.07ADD95 pKa = 2.99IAQRR99 pKa = 11.84MQAQSDD105 pKa = 4.27EE106 pKa = 4.39YY107 pKa = 11.43DD108 pKa = 3.61LVAVDD113 pKa = 3.64VTNTAEE119 pKa = 4.07FAANGWLLPLEE130 pKa = 4.51GDD132 pKa = 3.83NAIDD136 pKa = 3.9TDD138 pKa = 4.16GLLPATVEE146 pKa = 3.99SGTYY150 pKa = 10.44NGTLYY155 pKa = 10.8AAPKK159 pKa = 7.51NTNGAFLYY167 pKa = 10.7YY168 pKa = 9.71RR169 pKa = 11.84TDD171 pKa = 3.75LVDD174 pKa = 3.45QAPATWDD181 pKa = 3.43EE182 pKa = 4.75LKK184 pKa = 11.01AEE186 pKa = 4.28CPKK189 pKa = 10.96AEE191 pKa = 4.19EE192 pKa = 4.57AGIACYY198 pKa = 9.96IGQFSNYY205 pKa = 9.48EE206 pKa = 3.97GLTVNVTEE214 pKa = 4.55IVNAFGGSFVAEE226 pKa = 4.84DD227 pKa = 4.29GLTPAIDD234 pKa = 3.54EE235 pKa = 4.26QTAEE239 pKa = 4.14GLQFIVDD246 pKa = 3.93SFEE249 pKa = 5.64DD250 pKa = 3.87GTIPAAAQTYY260 pKa = 8.75TEE262 pKa = 5.06GEE264 pKa = 4.25SQTAFGNGEE273 pKa = 4.26ALFMRR278 pKa = 11.84TWPGFYY284 pKa = 9.75TDD286 pKa = 4.35GEE288 pKa = 4.45EE289 pKa = 4.0TSVAGNYY296 pKa = 9.63GIAVLPGVDD305 pKa = 3.68GPGVSTLGGYY315 pKa = 9.4NVGISAFTDD324 pKa = 3.66APATARR330 pKa = 11.84DD331 pKa = 3.91FLEE334 pKa = 4.71FMQTRR339 pKa = 11.84QSQEE343 pKa = 3.6YY344 pKa = 7.74YY345 pKa = 10.87AEE347 pKa = 4.22VGGAPVLAEE356 pKa = 4.27VYY358 pKa = 10.24DD359 pKa = 4.1DD360 pKa = 4.79PEE362 pKa = 4.43ILEE365 pKa = 4.08QYY367 pKa = 10.45PYY369 pKa = 11.08FSTLKK374 pKa = 10.32EE375 pKa = 3.98ALEE378 pKa = 4.35SAKK381 pKa = 10.02PRR383 pKa = 11.84PSSPYY388 pKa = 8.62YY389 pKa = 10.1SAVSKK394 pKa = 10.81AISDD398 pKa = 3.6NAYY401 pKa = 10.14AAIRR405 pKa = 11.84GEE407 pKa = 4.21KK408 pKa = 9.65PVQQALDD415 pKa = 3.62DD416 pKa = 4.03TQAGIEE422 pKa = 4.18TAGNN426 pKa = 3.39
Molecular weight: 45.06 kDa
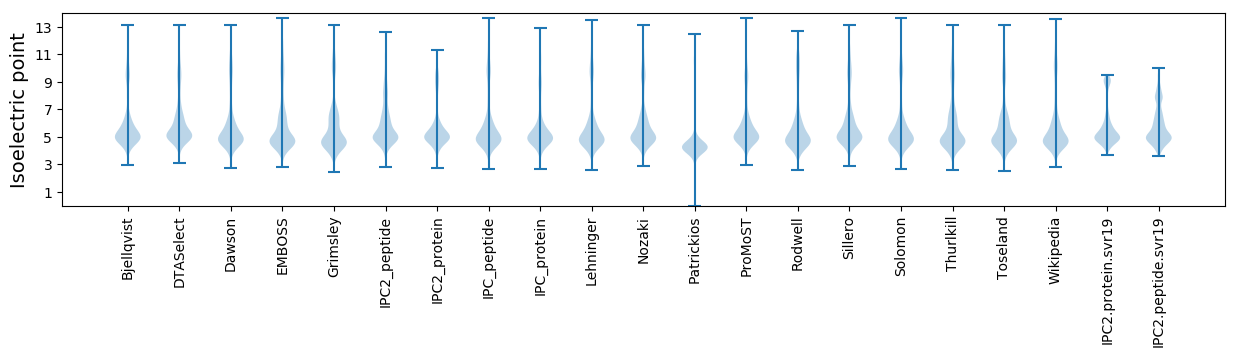
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R2D2Y9|A0A4R2D2Y9_9NOCA Uncharacterized protein YlxW (UPF0749 family) OS=Rhodococcus sp. SMB37 OX=2512213 GN=EV641_11866 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.64GRR42 pKa = 11.84ASLTAA47 pKa = 4.1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.64GRR42 pKa = 11.84ASLTAA47 pKa = 4.1
Molecular weight: 5.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1883713 |
26 |
9052 |
322.6 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.426 ± 0.045 | 0.763 ± 0.01 |
6.621 ± 0.032 | 5.758 ± 0.026 |
3.014 ± 0.017 | 8.816 ± 0.034 |
2.285 ± 0.018 | 4.297 ± 0.023 |
2.014 ± 0.026 | 9.865 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.98 ± 0.016 | 2.024 ± 0.017 |
5.57 ± 0.026 | 2.68 ± 0.016 |
7.372 ± 0.038 | 5.714 ± 0.021 |
6.387 ± 0.022 | 8.926 ± 0.036 |
1.45 ± 0.013 | 2.037 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
