
Wuhan Louse Fly Virus 10
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Lousefly sigmavirus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
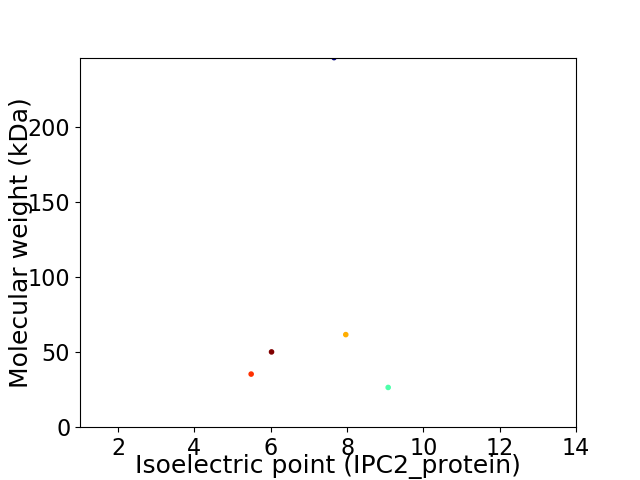
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KKC0|A0A0B5KKC0_9RHAB Matrix protein OS=Wuhan Louse Fly Virus 10 OX=1608114 GN=M PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.31RR3 pKa = 11.84RR4 pKa = 11.84MSSNEE9 pKa = 3.83LKK11 pKa = 10.59DD12 pKa = 3.92LRR14 pKa = 11.84AGTEE18 pKa = 3.45WDD20 pKa = 3.78LPASKK25 pKa = 10.22LRR27 pKa = 11.84KK28 pKa = 8.82LQEE31 pKa = 4.13AGVLNRR37 pKa = 11.84IYY39 pKa = 10.15EE40 pKa = 4.09QQQTSTIDD48 pKa = 3.26PTIGLDD54 pKa = 3.6PSIMEE59 pKa = 4.31HH60 pKa = 7.54DD61 pKa = 4.19YY62 pKa = 11.83DD63 pKa = 4.23EE64 pKa = 4.79TDD66 pKa = 3.08HH67 pKa = 6.53TLSLGDD73 pKa = 3.72SLMRR77 pKa = 11.84SPDD80 pKa = 3.51YY81 pKa = 10.83LIPPEE86 pKa = 4.43DD87 pKa = 3.83KK88 pKa = 10.98EE89 pKa = 4.3EE90 pKa = 4.14EE91 pKa = 4.21EE92 pKa = 4.51TLSEE96 pKa = 4.19TSRR99 pKa = 11.84EE100 pKa = 4.19DD101 pKa = 3.46EE102 pKa = 4.25NSAKK106 pKa = 10.49YY107 pKa = 10.29PEE109 pKa = 4.57FEE111 pKa = 4.15GSVMEE116 pKa = 4.37TFQIDD121 pKa = 3.85VTSPHH126 pKa = 6.66HH127 pKa = 6.9LSDD130 pKa = 5.05PEE132 pKa = 4.19LLCRR136 pKa = 11.84KK137 pKa = 7.93LTHH140 pKa = 7.25LIKK143 pKa = 10.77LVGNGRR149 pKa = 11.84IKK151 pKa = 10.41PEE153 pKa = 3.71QVKK156 pKa = 9.36FVHH159 pKa = 6.64KK160 pKa = 10.63NVEE163 pKa = 4.04KK164 pKa = 10.45PNTKK168 pKa = 9.72IIQDD172 pKa = 3.78KK173 pKa = 10.7GDD175 pKa = 3.53NTIDD179 pKa = 3.48SLEE182 pKa = 4.21AKK184 pKa = 10.17EE185 pKa = 4.64NLTLNEE191 pKa = 3.92DD192 pKa = 3.48HH193 pKa = 7.24KK194 pKa = 10.29EE195 pKa = 4.07TRR197 pKa = 11.84EE198 pKa = 3.68RR199 pKa = 11.84SAQSKK204 pKa = 9.86RR205 pKa = 11.84ASQKK209 pKa = 9.88VDD211 pKa = 3.15NDD213 pKa = 3.17EE214 pKa = 4.39KK215 pKa = 11.0LYY217 pKa = 10.57EE218 pKa = 3.95SHH220 pKa = 7.44PFIKK224 pKa = 10.31RR225 pKa = 11.84IADD228 pKa = 3.62GVEE231 pKa = 3.81IPHH234 pKa = 6.72ILNPKK239 pKa = 8.35RR240 pKa = 11.84TLKK243 pKa = 9.93IDD245 pKa = 3.46KK246 pKa = 8.49EE247 pKa = 4.33HH248 pKa = 7.46PDD250 pKa = 3.42FNLIRR255 pKa = 11.84IKK257 pKa = 10.35EE258 pKa = 4.24VYY260 pKa = 9.1PKK262 pKa = 10.31YY263 pKa = 10.46HH264 pKa = 6.13QFPKK268 pKa = 10.76DD269 pKa = 3.22FAKK272 pKa = 10.63FLITIAQMSKK282 pKa = 9.54KK283 pKa = 8.42WDD285 pKa = 4.24FYY287 pKa = 10.57RR288 pKa = 11.84RR289 pKa = 11.84TYY291 pKa = 11.01NWVLFAEE298 pKa = 4.78SGMEE302 pKa = 3.97HH303 pKa = 6.93
MM1 pKa = 7.35KK2 pKa = 10.31RR3 pKa = 11.84RR4 pKa = 11.84MSSNEE9 pKa = 3.83LKK11 pKa = 10.59DD12 pKa = 3.92LRR14 pKa = 11.84AGTEE18 pKa = 3.45WDD20 pKa = 3.78LPASKK25 pKa = 10.22LRR27 pKa = 11.84KK28 pKa = 8.82LQEE31 pKa = 4.13AGVLNRR37 pKa = 11.84IYY39 pKa = 10.15EE40 pKa = 4.09QQQTSTIDD48 pKa = 3.26PTIGLDD54 pKa = 3.6PSIMEE59 pKa = 4.31HH60 pKa = 7.54DD61 pKa = 4.19YY62 pKa = 11.83DD63 pKa = 4.23EE64 pKa = 4.79TDD66 pKa = 3.08HH67 pKa = 6.53TLSLGDD73 pKa = 3.72SLMRR77 pKa = 11.84SPDD80 pKa = 3.51YY81 pKa = 10.83LIPPEE86 pKa = 4.43DD87 pKa = 3.83KK88 pKa = 10.98EE89 pKa = 4.3EE90 pKa = 4.14EE91 pKa = 4.21EE92 pKa = 4.51TLSEE96 pKa = 4.19TSRR99 pKa = 11.84EE100 pKa = 4.19DD101 pKa = 3.46EE102 pKa = 4.25NSAKK106 pKa = 10.49YY107 pKa = 10.29PEE109 pKa = 4.57FEE111 pKa = 4.15GSVMEE116 pKa = 4.37TFQIDD121 pKa = 3.85VTSPHH126 pKa = 6.66HH127 pKa = 6.9LSDD130 pKa = 5.05PEE132 pKa = 4.19LLCRR136 pKa = 11.84KK137 pKa = 7.93LTHH140 pKa = 7.25LIKK143 pKa = 10.77LVGNGRR149 pKa = 11.84IKK151 pKa = 10.41PEE153 pKa = 3.71QVKK156 pKa = 9.36FVHH159 pKa = 6.64KK160 pKa = 10.63NVEE163 pKa = 4.04KK164 pKa = 10.45PNTKK168 pKa = 9.72IIQDD172 pKa = 3.78KK173 pKa = 10.7GDD175 pKa = 3.53NTIDD179 pKa = 3.48SLEE182 pKa = 4.21AKK184 pKa = 10.17EE185 pKa = 4.64NLTLNEE191 pKa = 3.92DD192 pKa = 3.48HH193 pKa = 7.24KK194 pKa = 10.29EE195 pKa = 4.07TRR197 pKa = 11.84EE198 pKa = 3.68RR199 pKa = 11.84SAQSKK204 pKa = 9.86RR205 pKa = 11.84ASQKK209 pKa = 9.88VDD211 pKa = 3.15NDD213 pKa = 3.17EE214 pKa = 4.39KK215 pKa = 11.0LYY217 pKa = 10.57EE218 pKa = 3.95SHH220 pKa = 7.44PFIKK224 pKa = 10.31RR225 pKa = 11.84IADD228 pKa = 3.62GVEE231 pKa = 3.81IPHH234 pKa = 6.72ILNPKK239 pKa = 8.35RR240 pKa = 11.84TLKK243 pKa = 9.93IDD245 pKa = 3.46KK246 pKa = 8.49EE247 pKa = 4.33HH248 pKa = 7.46PDD250 pKa = 3.42FNLIRR255 pKa = 11.84IKK257 pKa = 10.35EE258 pKa = 4.24VYY260 pKa = 9.1PKK262 pKa = 10.31YY263 pKa = 10.46HH264 pKa = 6.13QFPKK268 pKa = 10.76DD269 pKa = 3.22FAKK272 pKa = 10.63FLITIAQMSKK282 pKa = 9.54KK283 pKa = 8.42WDD285 pKa = 4.24FYY287 pKa = 10.57RR288 pKa = 11.84RR289 pKa = 11.84TYY291 pKa = 11.01NWVLFAEE298 pKa = 4.78SGMEE302 pKa = 3.97HH303 pKa = 6.93
Molecular weight: 35.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRT4|A0A0B5KRT4_9RHAB Glycoprotein OS=Wuhan Louse Fly Virus 10 OX=1608114 GN=G PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.51NMKK5 pKa = 9.71KK6 pKa = 10.41SNRR9 pKa = 11.84GYY11 pKa = 10.21IEE13 pKa = 3.78MNRR16 pKa = 11.84LRR18 pKa = 11.84KK19 pKa = 9.35IPPKK23 pKa = 10.22LRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.94NGKK30 pKa = 9.53PSSSTGAAHH39 pKa = 6.89TSQVSDD45 pKa = 3.12SSQWTTVHH53 pKa = 5.42FTVPEE58 pKa = 4.23GTGSSRR64 pKa = 11.84RR65 pKa = 11.84VSLIKK70 pKa = 10.89GNWTVTGTLRR80 pKa = 11.84VSCDD84 pKa = 3.13KK85 pKa = 11.25NLTNWSMMEE94 pKa = 3.79RR95 pKa = 11.84TLEE98 pKa = 4.67HH99 pKa = 6.88ILDD102 pKa = 4.03VYY104 pKa = 10.4DD105 pKa = 3.79GNMVFKK111 pKa = 10.42PLILLFYY118 pKa = 9.84WILGVNLAPIPGPSNRR134 pKa = 11.84HH135 pKa = 4.55CWGVDD140 pKa = 3.65FGEE143 pKa = 4.86PISLNHH149 pKa = 6.93RR150 pKa = 11.84IEE152 pKa = 5.31LIGSQDD158 pKa = 3.01FGYY161 pKa = 10.82SKK163 pKa = 11.1NFASNFRR170 pKa = 11.84GHH172 pKa = 6.83KK173 pKa = 8.74FTVEE177 pKa = 3.37FNINFVHH184 pKa = 5.48TQRR187 pKa = 11.84RR188 pKa = 11.84TTSVVTIVNSQVPHH202 pKa = 5.87FKK204 pKa = 10.14DD205 pKa = 3.04RR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84FRR210 pKa = 11.84EE211 pKa = 4.04LLEE214 pKa = 4.46GSQISYY220 pKa = 10.56EE221 pKa = 4.12YY222 pKa = 10.3TEE224 pKa = 4.39EE225 pKa = 4.1EE226 pKa = 4.58GLVLTT231 pKa = 5.33
MM1 pKa = 7.73KK2 pKa = 10.51NMKK5 pKa = 9.71KK6 pKa = 10.41SNRR9 pKa = 11.84GYY11 pKa = 10.21IEE13 pKa = 3.78MNRR16 pKa = 11.84LRR18 pKa = 11.84KK19 pKa = 9.35IPPKK23 pKa = 10.22LRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.94NGKK30 pKa = 9.53PSSSTGAAHH39 pKa = 6.89TSQVSDD45 pKa = 3.12SSQWTTVHH53 pKa = 5.42FTVPEE58 pKa = 4.23GTGSSRR64 pKa = 11.84RR65 pKa = 11.84VSLIKK70 pKa = 10.89GNWTVTGTLRR80 pKa = 11.84VSCDD84 pKa = 3.13KK85 pKa = 11.25NLTNWSMMEE94 pKa = 3.79RR95 pKa = 11.84TLEE98 pKa = 4.67HH99 pKa = 6.88ILDD102 pKa = 4.03VYY104 pKa = 10.4DD105 pKa = 3.79GNMVFKK111 pKa = 10.42PLILLFYY118 pKa = 9.84WILGVNLAPIPGPSNRR134 pKa = 11.84HH135 pKa = 4.55CWGVDD140 pKa = 3.65FGEE143 pKa = 4.86PISLNHH149 pKa = 6.93RR150 pKa = 11.84IEE152 pKa = 5.31LIGSQDD158 pKa = 3.01FGYY161 pKa = 10.82SKK163 pKa = 11.1NFASNFRR170 pKa = 11.84GHH172 pKa = 6.83KK173 pKa = 8.74FTVEE177 pKa = 3.37FNINFVHH184 pKa = 5.48TQRR187 pKa = 11.84RR188 pKa = 11.84TTSVVTIVNSQVPHH202 pKa = 5.87FKK204 pKa = 10.14DD205 pKa = 3.04RR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84FRR210 pKa = 11.84EE211 pKa = 4.04LLEE214 pKa = 4.46GSQISYY220 pKa = 10.56EE221 pKa = 4.12YY222 pKa = 10.3TEE224 pKa = 4.39EE225 pKa = 4.1EE226 pKa = 4.58GLVLTT231 pKa = 5.33
Molecular weight: 26.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3661 |
231 |
2146 |
732.2 |
84.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.851 ± 0.812 | 1.475 ± 0.309 |
4.725 ± 0.495 | 6.501 ± 0.646 |
4.562 ± 0.259 | 5.299 ± 0.465 |
3.114 ± 0.165 | 8.468 ± 0.631 |
7.02 ± 0.619 | 10.653 ± 0.788 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.021 ± 0.325 | 5.272 ± 0.48 |
4.753 ± 0.301 | 3.305 ± 0.218 |
4.807 ± 0.613 | 8.14 ± 0.28 |
5.763 ± 0.473 | 4.835 ± 0.345 |
1.694 ± 0.144 | 3.742 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
