
Oikopleura dioica (Tunicate)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Tunicata; Appendicularia; Copelata; Oikopleuridae; Oikopleura
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
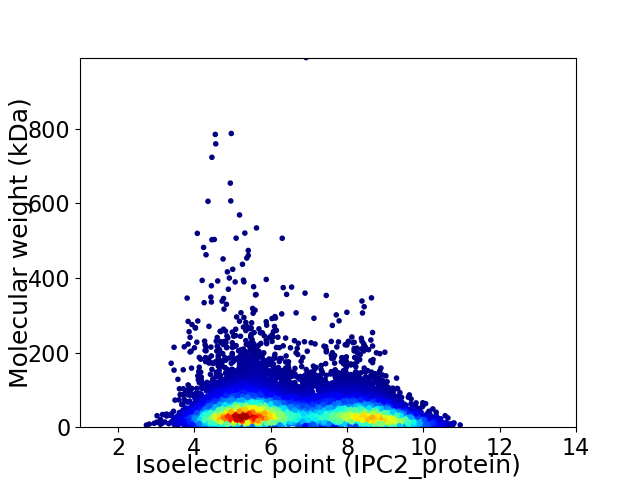
Virtual 2D-PAGE plot for 17050 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4Y370|E4Y370_OIKDI Uncharacterized protein (Fragment) OS=Oikopleura dioica OX=34765 GN=GSOID_T00001413001 PE=4 SV=1
CC1 pKa = 7.15HH2 pKa = 7.1AFLASRR8 pKa = 11.84ALTHH12 pKa = 6.5FLAPFVATAQLAFSEE27 pKa = 4.69TDD29 pKa = 3.38TIAQKK34 pKa = 9.46TKK36 pKa = 10.74SSMKK40 pKa = 10.14IITTTTITTTNISRR54 pKa = 11.84TNATSAGQDD63 pKa = 3.21AQTEE67 pKa = 4.48LVMIAQMDD75 pKa = 4.48LLEE78 pKa = 4.59MEE80 pKa = 4.5STVLILMNVKK90 pKa = 10.38KK91 pKa = 10.61EE92 pKa = 4.23GEE94 pKa = 4.17CVEE97 pKa = 4.6NSEE100 pKa = 4.81CLNFAGGFKK109 pKa = 10.52CGEE112 pKa = 4.33CEE114 pKa = 5.36LGFTGNQTVGCFSYY128 pKa = 11.35CNGCAEE134 pKa = 4.17NAICEE139 pKa = 4.28FAEE142 pKa = 4.32DD143 pKa = 3.79TWSHH147 pKa = 6.07FCSCKK152 pKa = 10.16TGFAGNGKK160 pKa = 8.98EE161 pKa = 4.67DD162 pKa = 3.83CGKK165 pKa = 9.06DD166 pKa = 3.06TDD168 pKa = 5.39LDD170 pKa = 3.99GFPDD174 pKa = 4.06DD175 pKa = 4.71VLNCRR180 pKa = 11.84DD181 pKa = 3.55SSCRR185 pKa = 11.84ADD187 pKa = 3.45NCATIPNSGQEE198 pKa = 4.43DD199 pKa = 3.99SDD201 pKa = 4.09SDD203 pKa = 4.68GIGDD207 pKa = 4.5ACDD210 pKa = 3.59NDD212 pKa = 3.45IDD214 pKa = 4.63NDD216 pKa = 4.76GITNVNDD223 pKa = 3.28NCPRR227 pKa = 11.84LANPSQVDD235 pKa = 3.64DD236 pKa = 5.38DD237 pKa = 5.0DD238 pKa = 7.08DD239 pKa = 4.84GVGDD243 pKa = 4.18EE244 pKa = 5.41CDD246 pKa = 3.53NCPTIYY252 pKa = 10.68NPSQDD257 pKa = 3.72DD258 pKa = 3.71SCIVMDD264 pKa = 4.65SDD266 pKa = 4.49GDD268 pKa = 4.32GISDD272 pKa = 3.59EE273 pKa = 5.38DD274 pKa = 4.63DD275 pKa = 3.46NCVEE279 pKa = 4.97IPNPDD284 pKa = 3.51QLDD287 pKa = 3.57FDD289 pKa = 5.43LDD291 pKa = 4.04GLGDD295 pKa = 3.63ICDD298 pKa = 4.05VDD300 pKa = 5.54DD301 pKa = 6.59DD302 pKa = 5.36DD303 pKa = 7.53DD304 pKa = 4.58GVPDD308 pKa = 5.71DD309 pKa = 5.58VDD311 pKa = 3.77DD312 pKa = 4.56CPRR315 pKa = 11.84VYY317 pKa = 10.79NPEE320 pKa = 3.92QLGCGPDD327 pKa = 3.44SDD329 pKa = 5.69GDD331 pKa = 3.92GTPDD335 pKa = 4.52EE336 pKa = 5.74FDD338 pKa = 3.24ICPNDD343 pKa = 3.91ALISSPSFTGYY354 pKa = 8.14EE355 pKa = 4.24TILLDD360 pKa = 4.55PEE362 pKa = 5.19GSSQLDD368 pKa = 3.96PYY370 pKa = 10.39WIVLNEE376 pKa = 3.75GRR378 pKa = 11.84EE379 pKa = 4.33LIQSVNADD387 pKa = 3.45PGLAIGKK394 pKa = 9.4HH395 pKa = 4.57NVQGVDD401 pKa = 3.42YY402 pKa = 10.74SGTVYY407 pKa = 10.7INTDD411 pKa = 2.97TDD413 pKa = 3.82DD414 pKa = 4.71DD415 pKa = 4.27FVGFVFGFQSSSRR428 pKa = 11.84FYY430 pKa = 11.02SVMWKK435 pKa = 9.57QVEE438 pKa = 4.01QTYY441 pKa = 8.43WWPSPFRR448 pKa = 11.84SVAPTGLQIKK458 pKa = 9.9LVDD461 pKa = 3.78SATGPGPLLRR471 pKa = 11.84NALWHH476 pKa = 6.87GDD478 pKa = 3.67DD479 pKa = 3.87TPGQVKK485 pKa = 9.07VLWEE489 pKa = 4.43DD490 pKa = 3.47EE491 pKa = 4.38NKK493 pKa = 10.38IGWTDD498 pKa = 3.61FTAYY502 pKa = 9.89RR503 pKa = 11.84WVLHH507 pKa = 5.94HH508 pKa = 6.95RR509 pKa = 11.84PSTGLIQVQIFEE521 pKa = 4.85GEE523 pKa = 4.2TKK525 pKa = 10.23IIDD528 pKa = 3.73SGNVIDD534 pKa = 4.18TTFQGGRR541 pKa = 11.84LGVLDD546 pKa = 4.84FSQEE550 pKa = 3.76NVIWSNLNYY559 pKa = 10.13RR560 pKa = 11.84CNEE563 pKa = 3.89TMPLFF568 pKa = 4.18
CC1 pKa = 7.15HH2 pKa = 7.1AFLASRR8 pKa = 11.84ALTHH12 pKa = 6.5FLAPFVATAQLAFSEE27 pKa = 4.69TDD29 pKa = 3.38TIAQKK34 pKa = 9.46TKK36 pKa = 10.74SSMKK40 pKa = 10.14IITTTTITTTNISRR54 pKa = 11.84TNATSAGQDD63 pKa = 3.21AQTEE67 pKa = 4.48LVMIAQMDD75 pKa = 4.48LLEE78 pKa = 4.59MEE80 pKa = 4.5STVLILMNVKK90 pKa = 10.38KK91 pKa = 10.61EE92 pKa = 4.23GEE94 pKa = 4.17CVEE97 pKa = 4.6NSEE100 pKa = 4.81CLNFAGGFKK109 pKa = 10.52CGEE112 pKa = 4.33CEE114 pKa = 5.36LGFTGNQTVGCFSYY128 pKa = 11.35CNGCAEE134 pKa = 4.17NAICEE139 pKa = 4.28FAEE142 pKa = 4.32DD143 pKa = 3.79TWSHH147 pKa = 6.07FCSCKK152 pKa = 10.16TGFAGNGKK160 pKa = 8.98EE161 pKa = 4.67DD162 pKa = 3.83CGKK165 pKa = 9.06DD166 pKa = 3.06TDD168 pKa = 5.39LDD170 pKa = 3.99GFPDD174 pKa = 4.06DD175 pKa = 4.71VLNCRR180 pKa = 11.84DD181 pKa = 3.55SSCRR185 pKa = 11.84ADD187 pKa = 3.45NCATIPNSGQEE198 pKa = 4.43DD199 pKa = 3.99SDD201 pKa = 4.09SDD203 pKa = 4.68GIGDD207 pKa = 4.5ACDD210 pKa = 3.59NDD212 pKa = 3.45IDD214 pKa = 4.63NDD216 pKa = 4.76GITNVNDD223 pKa = 3.28NCPRR227 pKa = 11.84LANPSQVDD235 pKa = 3.64DD236 pKa = 5.38DD237 pKa = 5.0DD238 pKa = 7.08DD239 pKa = 4.84GVGDD243 pKa = 4.18EE244 pKa = 5.41CDD246 pKa = 3.53NCPTIYY252 pKa = 10.68NPSQDD257 pKa = 3.72DD258 pKa = 3.71SCIVMDD264 pKa = 4.65SDD266 pKa = 4.49GDD268 pKa = 4.32GISDD272 pKa = 3.59EE273 pKa = 5.38DD274 pKa = 4.63DD275 pKa = 3.46NCVEE279 pKa = 4.97IPNPDD284 pKa = 3.51QLDD287 pKa = 3.57FDD289 pKa = 5.43LDD291 pKa = 4.04GLGDD295 pKa = 3.63ICDD298 pKa = 4.05VDD300 pKa = 5.54DD301 pKa = 6.59DD302 pKa = 5.36DD303 pKa = 7.53DD304 pKa = 4.58GVPDD308 pKa = 5.71DD309 pKa = 5.58VDD311 pKa = 3.77DD312 pKa = 4.56CPRR315 pKa = 11.84VYY317 pKa = 10.79NPEE320 pKa = 3.92QLGCGPDD327 pKa = 3.44SDD329 pKa = 5.69GDD331 pKa = 3.92GTPDD335 pKa = 4.52EE336 pKa = 5.74FDD338 pKa = 3.24ICPNDD343 pKa = 3.91ALISSPSFTGYY354 pKa = 8.14EE355 pKa = 4.24TILLDD360 pKa = 4.55PEE362 pKa = 5.19GSSQLDD368 pKa = 3.96PYY370 pKa = 10.39WIVLNEE376 pKa = 3.75GRR378 pKa = 11.84EE379 pKa = 4.33LIQSVNADD387 pKa = 3.45PGLAIGKK394 pKa = 9.4HH395 pKa = 4.57NVQGVDD401 pKa = 3.42YY402 pKa = 10.74SGTVYY407 pKa = 10.7INTDD411 pKa = 2.97TDD413 pKa = 3.82DD414 pKa = 4.71DD415 pKa = 4.27FVGFVFGFQSSSRR428 pKa = 11.84FYY430 pKa = 11.02SVMWKK435 pKa = 9.57QVEE438 pKa = 4.01QTYY441 pKa = 8.43WWPSPFRR448 pKa = 11.84SVAPTGLQIKK458 pKa = 9.9LVDD461 pKa = 3.78SATGPGPLLRR471 pKa = 11.84NALWHH476 pKa = 6.87GDD478 pKa = 3.67DD479 pKa = 3.87TPGQVKK485 pKa = 9.07VLWEE489 pKa = 4.43DD490 pKa = 3.47EE491 pKa = 4.38NKK493 pKa = 10.38IGWTDD498 pKa = 3.61FTAYY502 pKa = 9.89RR503 pKa = 11.84WVLHH507 pKa = 5.94HH508 pKa = 6.95RR509 pKa = 11.84PSTGLIQVQIFEE521 pKa = 4.85GEE523 pKa = 4.2TKK525 pKa = 10.23IIDD528 pKa = 3.73SGNVIDD534 pKa = 4.18TTFQGGRR541 pKa = 11.84LGVLDD546 pKa = 4.84FSQEE550 pKa = 3.76NVIWSNLNYY559 pKa = 10.13RR560 pKa = 11.84CNEE563 pKa = 3.89TMPLFF568 pKa = 4.18
Molecular weight: 61.92 kDa
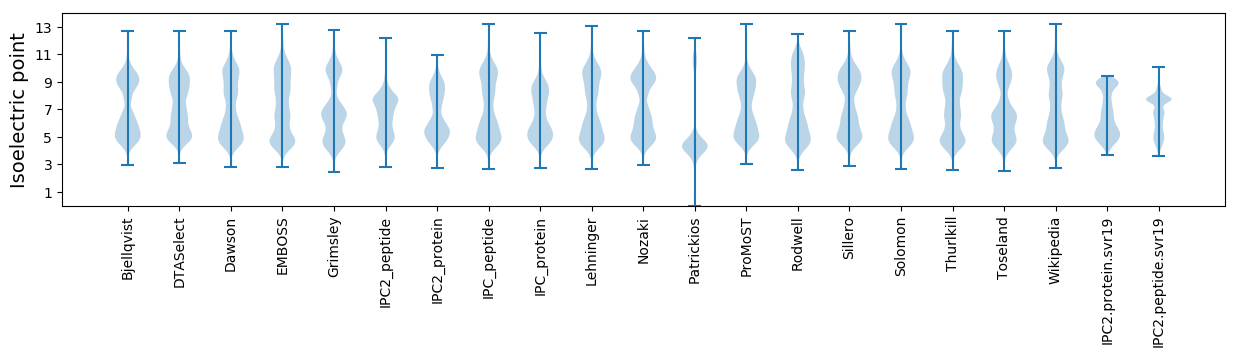
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4X731|E4X731_OIKDI Septin OS=Oikopleura dioica OX=34765 GN=GSOID_T00003368001 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 10.19ISTRR6 pKa = 11.84SRR8 pKa = 11.84TTSTKK13 pKa = 10.8LPMQVLSTRR22 pKa = 11.84IQAASSRR29 pKa = 11.84SAITRR34 pKa = 11.84KK35 pKa = 9.7PPNTLVLIMTKK46 pKa = 9.79TSGFAPISRR55 pKa = 4.08
MM1 pKa = 7.54KK2 pKa = 10.19ISTRR6 pKa = 11.84SRR8 pKa = 11.84TTSTKK13 pKa = 10.8LPMQVLSTRR22 pKa = 11.84IQAASSRR29 pKa = 11.84SAITRR34 pKa = 11.84KK35 pKa = 9.7PPNTLVLIMTKK46 pKa = 9.79TSGFAPISRR55 pKa = 4.08
Molecular weight: 6.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6837326 |
10 |
8885 |
401.0 |
44.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.446 ± 0.021 | 2.1 ± 0.026 |
5.765 ± 0.016 | 7.396 ± 0.024 |
4.46 ± 0.017 | 5.444 ± 0.024 |
2.032 ± 0.008 | 5.829 ± 0.017 |
6.953 ± 0.03 | 8.333 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.265 ± 0.009 | 4.973 ± 0.014 |
4.454 ± 0.019 | 3.982 ± 0.017 |
5.31 ± 0.02 | 7.924 ± 0.023 |
5.403 ± 0.023 | 5.441 ± 0.017 |
1.182 ± 0.007 | 2.846 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
