
Lesser panda anellovirus
Taxonomy: Viruses; Anelloviridae; Thetatorquevirus; Torque teno ursid virus 1
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
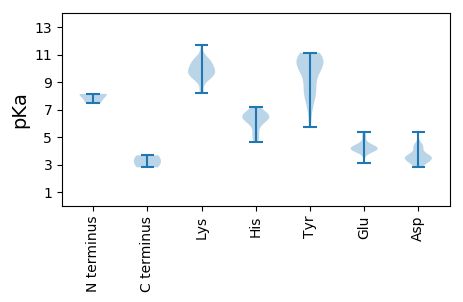
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2Z2L0X1|A0A2Z2L0X1_9VIRU Capsid protein OS=Lesser panda anellovirus OX=2012640 PE=3 SV=1
MM1 pKa = 8.14VEE3 pKa = 5.05FILPYY8 pKa = 8.34GQSRR12 pKa = 11.84RR13 pKa = 11.84SCKK16 pKa = 9.79RR17 pKa = 11.84SRR19 pKa = 11.84FDD21 pKa = 3.68AVGGCRR27 pKa = 11.84RR28 pKa = 11.84TSGACPEE35 pKa = 4.04SRR37 pKa = 11.84GLSGKK42 pKa = 10.47GEE44 pKa = 4.0YY45 pKa = 9.95RR46 pKa = 11.84QMGSPEE52 pKa = 4.2GQSADD57 pKa = 3.46AGHH60 pKa = 6.58SEE62 pKa = 4.22TLWLEE67 pKa = 4.17SCLLTHH73 pKa = 7.18SLWCKK78 pKa = 9.29CNDD81 pKa = 2.83WRR83 pKa = 11.84EE84 pKa = 4.27HH85 pKa = 4.64VRR87 pKa = 11.84KK88 pKa = 8.24TDD90 pKa = 3.47STWRR94 pKa = 11.84SSTGGGDD101 pKa = 3.17RR102 pKa = 11.84TGEE105 pKa = 3.95EE106 pKa = 4.21GGVPDD111 pKa = 4.26EE112 pKa = 4.52VAVHH116 pKa = 6.36FDD118 pKa = 3.47LGFSEE123 pKa = 5.38EE124 pKa = 4.47DD125 pKa = 3.22DD126 pKa = 3.68TRR128 pKa = 11.84TGEE131 pKa = 3.97RR132 pKa = 3.67
MM1 pKa = 8.14VEE3 pKa = 5.05FILPYY8 pKa = 8.34GQSRR12 pKa = 11.84RR13 pKa = 11.84SCKK16 pKa = 9.79RR17 pKa = 11.84SRR19 pKa = 11.84FDD21 pKa = 3.68AVGGCRR27 pKa = 11.84RR28 pKa = 11.84TSGACPEE35 pKa = 4.04SRR37 pKa = 11.84GLSGKK42 pKa = 10.47GEE44 pKa = 4.0YY45 pKa = 9.95RR46 pKa = 11.84QMGSPEE52 pKa = 4.2GQSADD57 pKa = 3.46AGHH60 pKa = 6.58SEE62 pKa = 4.22TLWLEE67 pKa = 4.17SCLLTHH73 pKa = 7.18SLWCKK78 pKa = 9.29CNDD81 pKa = 2.83WRR83 pKa = 11.84EE84 pKa = 4.27HH85 pKa = 4.64VRR87 pKa = 11.84KK88 pKa = 8.24TDD90 pKa = 3.47STWRR94 pKa = 11.84SSTGGGDD101 pKa = 3.17RR102 pKa = 11.84TGEE105 pKa = 3.95EE106 pKa = 4.21GGVPDD111 pKa = 4.26EE112 pKa = 4.52VAVHH116 pKa = 6.36FDD118 pKa = 3.47LGFSEE123 pKa = 5.38EE124 pKa = 4.47DD125 pKa = 3.22DD126 pKa = 3.68TRR128 pKa = 11.84TGEE131 pKa = 3.97RR132 pKa = 3.67
Molecular weight: 14.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z2L314|A0A2Z2L314_9VIRU ORF3 OS=Lesser panda anellovirus OX=2012640 PE=4 SV=1
MM1 pKa = 7.48AFFYY5 pKa = 10.77RR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84PYY11 pKa = 8.07WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84WCPRR19 pKa = 11.84RR20 pKa = 11.84GGRR23 pKa = 11.84SFRR26 pKa = 11.84PRR28 pKa = 11.84IFRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.02TYY37 pKa = 8.52RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.04VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84VRR54 pKa = 11.84PKK56 pKa = 10.48KK57 pKa = 9.97IIQWQPQHH65 pKa = 5.24QAKK68 pKa = 10.36CIIRR72 pKa = 11.84GWFPAMWAHH81 pKa = 6.13SAFVTYY87 pKa = 10.13PMYY90 pKa = 10.86KK91 pKa = 9.76WIPAKK96 pKa = 10.21QGEE99 pKa = 4.48YY100 pKa = 10.57GHH102 pKa = 6.68FFTMEE107 pKa = 3.84GCVTLMHH114 pKa = 6.36FTLGLLYY121 pKa = 10.57KK122 pKa = 10.12EE123 pKa = 4.07WLKK126 pKa = 10.19MRR128 pKa = 11.84NTWSRR133 pKa = 11.84SNDD136 pKa = 3.06GFDD139 pKa = 3.14LAKK142 pKa = 10.9YY143 pKa = 10.28SGTKK147 pKa = 8.84LTFRR151 pKa = 11.84PHH153 pKa = 6.56PFVSYY158 pKa = 8.32MVWWEE163 pKa = 3.99RR164 pKa = 11.84NFGEE168 pKa = 4.26VHH170 pKa = 6.92PIQYY174 pKa = 10.21RR175 pKa = 11.84SIHH178 pKa = 6.34PAVIMHH184 pKa = 5.96QPNHH188 pKa = 6.17IFVPSLLVNPXGRR201 pKa = 11.84KK202 pKa = 8.17RR203 pKa = 11.84KK204 pKa = 9.24VWIPPPSTLTSTWSFMKK221 pKa = 9.83YY222 pKa = 5.73WCEE225 pKa = 3.48VALFRR230 pKa = 11.84VGITPINLKK239 pKa = 9.97SQFLHH244 pKa = 6.51IGTVSTGWWIGYY256 pKa = 7.31QPKK259 pKa = 10.56NNYY262 pKa = 8.01NHH264 pKa = 6.6FSDD267 pKa = 5.37PDD269 pKa = 3.66MLPQMNSEE277 pKa = 4.28QYY279 pKa = 10.78CVGEE283 pKa = 4.24GNKK286 pKa = 9.33IFYY289 pKa = 9.2KK290 pKa = 9.77WWWDD294 pKa = 3.56DD295 pKa = 3.32GTDD298 pKa = 3.86NKK300 pKa = 10.1WGAKK304 pKa = 9.31NRR306 pKa = 11.84TDD308 pKa = 3.39QYY310 pKa = 11.13NNEE313 pKa = 4.34TGVLPPVAQKK323 pKa = 9.93TSTDD327 pKa = 3.47NIVPINVPYY336 pKa = 10.21WLAFWGAGTTFLEE349 pKa = 4.02WDD351 pKa = 4.02KK352 pKa = 11.66NDD354 pKa = 4.38LYY356 pKa = 10.79IWWYY360 pKa = 10.69YY361 pKa = 11.06DD362 pKa = 4.28DD363 pKa = 4.45NSKK366 pKa = 9.75TNIDD370 pKa = 4.17EE371 pKa = 4.71LDD373 pKa = 3.5GAAKK377 pKa = 9.47KK378 pKa = 10.34KK379 pKa = 9.95RR380 pKa = 11.84WINVSCGQMLNGIGIQNIIQGGPFVLNPNDD410 pKa = 3.6IFPEE414 pKa = 4.11KK415 pKa = 10.59NINLSFWYY423 pKa = 10.16QSFWRR428 pKa = 11.84WGGISPSPPINVDD441 pKa = 3.15PCQVDD446 pKa = 3.67PAHH449 pKa = 6.57APQRR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84VPRR458 pKa = 11.84GVPEE462 pKa = 4.28SPKK465 pKa = 10.71HH466 pKa = 5.83VGDD469 pKa = 3.84WSLHH473 pKa = 5.26PWDD476 pKa = 4.6LTRR479 pKa = 11.84QGLITEE485 pKa = 5.01AKK487 pKa = 9.44LKK489 pKa = 10.55QIFGFSPDD497 pKa = 4.48DD498 pKa = 3.5IAGLPHH504 pKa = 6.7PPSRR508 pKa = 11.84EE509 pKa = 3.97SSPATTDD516 pKa = 3.0SSPPPTQLRR525 pKa = 11.84AVRR528 pKa = 11.84RR529 pKa = 11.84KK530 pKa = 9.63RR531 pKa = 11.84SRR533 pKa = 11.84SEE535 pKa = 5.03DD536 pKa = 2.91LWQSPDD542 pKa = 3.39SEE544 pKa = 4.41EE545 pKa = 4.65EE546 pKa = 4.06KK547 pKa = 11.03VQTGDD552 pKa = 3.49TLSLLAQRR560 pKa = 11.84IEE562 pKa = 3.89RR563 pKa = 11.84EE564 pKa = 3.11RR565 pKa = 11.84DD566 pKa = 3.18FRR568 pKa = 11.84HH569 pKa = 6.55KK570 pKa = 10.12LRR572 pKa = 11.84KK573 pKa = 9.58RR574 pKa = 11.84IRR576 pKa = 11.84DD577 pKa = 3.31WLTT580 pKa = 2.8
MM1 pKa = 7.48AFFYY5 pKa = 10.77RR6 pKa = 11.84RR7 pKa = 11.84WRR9 pKa = 11.84PYY11 pKa = 8.07WRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84WCPRR19 pKa = 11.84RR20 pKa = 11.84GGRR23 pKa = 11.84SFRR26 pKa = 11.84PRR28 pKa = 11.84IFRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.02TYY37 pKa = 8.52RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.04VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84VRR54 pKa = 11.84PKK56 pKa = 10.48KK57 pKa = 9.97IIQWQPQHH65 pKa = 5.24QAKK68 pKa = 10.36CIIRR72 pKa = 11.84GWFPAMWAHH81 pKa = 6.13SAFVTYY87 pKa = 10.13PMYY90 pKa = 10.86KK91 pKa = 9.76WIPAKK96 pKa = 10.21QGEE99 pKa = 4.48YY100 pKa = 10.57GHH102 pKa = 6.68FFTMEE107 pKa = 3.84GCVTLMHH114 pKa = 6.36FTLGLLYY121 pKa = 10.57KK122 pKa = 10.12EE123 pKa = 4.07WLKK126 pKa = 10.19MRR128 pKa = 11.84NTWSRR133 pKa = 11.84SNDD136 pKa = 3.06GFDD139 pKa = 3.14LAKK142 pKa = 10.9YY143 pKa = 10.28SGTKK147 pKa = 8.84LTFRR151 pKa = 11.84PHH153 pKa = 6.56PFVSYY158 pKa = 8.32MVWWEE163 pKa = 3.99RR164 pKa = 11.84NFGEE168 pKa = 4.26VHH170 pKa = 6.92PIQYY174 pKa = 10.21RR175 pKa = 11.84SIHH178 pKa = 6.34PAVIMHH184 pKa = 5.96QPNHH188 pKa = 6.17IFVPSLLVNPXGRR201 pKa = 11.84KK202 pKa = 8.17RR203 pKa = 11.84KK204 pKa = 9.24VWIPPPSTLTSTWSFMKK221 pKa = 9.83YY222 pKa = 5.73WCEE225 pKa = 3.48VALFRR230 pKa = 11.84VGITPINLKK239 pKa = 9.97SQFLHH244 pKa = 6.51IGTVSTGWWIGYY256 pKa = 7.31QPKK259 pKa = 10.56NNYY262 pKa = 8.01NHH264 pKa = 6.6FSDD267 pKa = 5.37PDD269 pKa = 3.66MLPQMNSEE277 pKa = 4.28QYY279 pKa = 10.78CVGEE283 pKa = 4.24GNKK286 pKa = 9.33IFYY289 pKa = 9.2KK290 pKa = 9.77WWWDD294 pKa = 3.56DD295 pKa = 3.32GTDD298 pKa = 3.86NKK300 pKa = 10.1WGAKK304 pKa = 9.31NRR306 pKa = 11.84TDD308 pKa = 3.39QYY310 pKa = 11.13NNEE313 pKa = 4.34TGVLPPVAQKK323 pKa = 9.93TSTDD327 pKa = 3.47NIVPINVPYY336 pKa = 10.21WLAFWGAGTTFLEE349 pKa = 4.02WDD351 pKa = 4.02KK352 pKa = 11.66NDD354 pKa = 4.38LYY356 pKa = 10.79IWWYY360 pKa = 10.69YY361 pKa = 11.06DD362 pKa = 4.28DD363 pKa = 4.45NSKK366 pKa = 9.75TNIDD370 pKa = 4.17EE371 pKa = 4.71LDD373 pKa = 3.5GAAKK377 pKa = 9.47KK378 pKa = 10.34KK379 pKa = 9.95RR380 pKa = 11.84WINVSCGQMLNGIGIQNIIQGGPFVLNPNDD410 pKa = 3.6IFPEE414 pKa = 4.11KK415 pKa = 10.59NINLSFWYY423 pKa = 10.16QSFWRR428 pKa = 11.84WGGISPSPPINVDD441 pKa = 3.15PCQVDD446 pKa = 3.67PAHH449 pKa = 6.57APQRR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84VPRR458 pKa = 11.84GVPEE462 pKa = 4.28SPKK465 pKa = 10.71HH466 pKa = 5.83VGDD469 pKa = 3.84WSLHH473 pKa = 5.26PWDD476 pKa = 4.6LTRR479 pKa = 11.84QGLITEE485 pKa = 5.01AKK487 pKa = 9.44LKK489 pKa = 10.55QIFGFSPDD497 pKa = 4.48DD498 pKa = 3.5IAGLPHH504 pKa = 6.7PPSRR508 pKa = 11.84EE509 pKa = 3.97SSPATTDD516 pKa = 3.0SSPPPTQLRR525 pKa = 11.84AVRR528 pKa = 11.84RR529 pKa = 11.84KK530 pKa = 9.63RR531 pKa = 11.84SRR533 pKa = 11.84SEE535 pKa = 5.03DD536 pKa = 2.91LWQSPDD542 pKa = 3.39SEE544 pKa = 4.41EE545 pKa = 4.65EE546 pKa = 4.06KK547 pKa = 11.03VQTGDD552 pKa = 3.49TLSLLAQRR560 pKa = 11.84IEE562 pKa = 3.89RR563 pKa = 11.84EE564 pKa = 3.11RR565 pKa = 11.84DD566 pKa = 3.18FRR568 pKa = 11.84HH569 pKa = 6.55KK570 pKa = 10.12LRR572 pKa = 11.84KK573 pKa = 9.58RR574 pKa = 11.84IRR576 pKa = 11.84DD577 pKa = 3.31WLTT580 pKa = 2.8
Molecular weight: 68.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974 |
132 |
580 |
324.7 |
37.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.209 ± 0.449 | 2.156 ± 0.919 |
4.928 ± 0.533 | 5.749 ± 1.933 |
3.799 ± 0.909 | 8.316 ± 2.045 |
2.669 ± 0.157 | 3.799 ± 1.821 |
5.031 ± 0.687 | 5.955 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.745 ± 0.286 | 3.388 ± 1.297 |
6.982 ± 1.455 | 3.491 ± 0.724 |
10.575 ± 0.535 | 9.035 ± 2.647 |
6.263 ± 0.976 | 4.415 ± 0.402 |
4.312 ± 1.21 | 3.08 ± 0.665 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
