
Acidianus filamentous virus 6
Taxonomy: Viruses; Adnaviria; Zilligvirae; Taleaviricota; Tokiviricetes; Ligamenvirales; Lipothrixviridae; Betalipothrixvirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
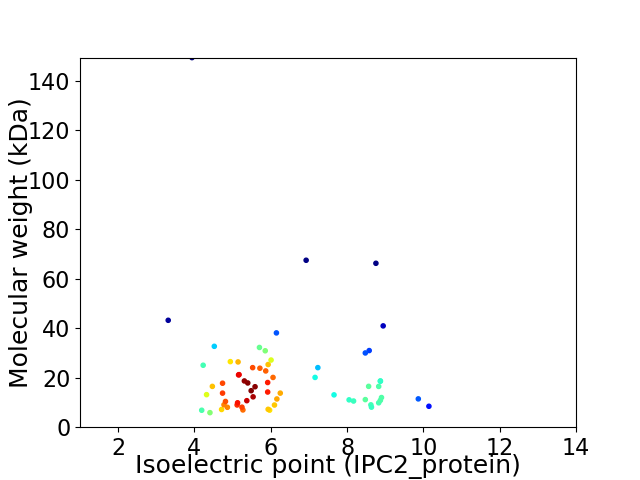
Virtual 2D-PAGE plot for 66 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7WKL0|A7WKL0_9VIRU Uncharacterized protein OS=Acidianus filamentous virus 6 OX=346882 PE=4 SV=1
MM1 pKa = 7.68SCGEE5 pKa = 4.8LPNASCTPTPNWEE18 pKa = 4.25CSDD21 pKa = 3.8VEE23 pKa = 5.31CFQQCINQPTFEE35 pKa = 4.63QFYY38 pKa = 9.5EE39 pKa = 4.38CCIQCALAHH48 pKa = 6.14YY49 pKa = 9.62IAEE52 pKa = 4.68AKK54 pKa = 10.57DD55 pKa = 3.31HH56 pKa = 5.05VTILVANAVSMIGTEE71 pKa = 4.18YY72 pKa = 10.6IPVFDD77 pKa = 3.95QMQMFPAHH85 pKa = 7.15LEE87 pKa = 3.98TLQGKK92 pKa = 7.47EE93 pKa = 3.77QVTVRR98 pKa = 11.84DD99 pKa = 3.4LVFPQNYY106 pKa = 8.08PVKK109 pKa = 10.5KK110 pKa = 7.46YY111 pKa = 8.92TYY113 pKa = 9.33YY114 pKa = 11.18EE115 pKa = 3.97NVQVNDD121 pKa = 3.06IGQPCSSPTGFCVPPGVTASEE142 pKa = 4.17EE143 pKa = 3.89LWFVNEE149 pKa = 4.03GSVVIIPSTLPPTTVSTFSYY169 pKa = 8.58ITPPPPISFQVTYY182 pKa = 7.7PTLVVPPPPPSPPPTTNILVVSKK205 pKa = 9.54PCPEE209 pKa = 3.95VCPGCICYY217 pKa = 10.68YY218 pKa = 9.68MLGIDD223 pKa = 4.76IVDD226 pKa = 3.85PTDD229 pKa = 3.12AVYY232 pKa = 10.61NYY234 pKa = 9.99QEE236 pKa = 4.3TPTGVVQQVPQNEE249 pKa = 4.36YY250 pKa = 10.98VLPSDD255 pKa = 4.12LLSIEE260 pKa = 4.42EE261 pKa = 4.58APSHH265 pKa = 4.59TTEE268 pKa = 4.11TEE270 pKa = 4.11TVEE273 pKa = 4.0EE274 pKa = 4.65SISFTLYY281 pKa = 10.59YY282 pKa = 10.73VVTTTLPVLDD292 pKa = 4.17VNFVYY297 pKa = 10.03PVTLIRR303 pKa = 11.84EE304 pKa = 4.47VEE306 pKa = 4.32SVTASRR312 pKa = 11.84SIQTVYY318 pKa = 10.48FSYY321 pKa = 10.35PVSKK325 pKa = 10.43QVFSEE330 pKa = 4.45YY331 pKa = 8.61VTASSISVSAYY342 pKa = 7.48FQPIVKK348 pKa = 9.84LVQEE352 pKa = 4.47SEE354 pKa = 4.6VTVSSIPILQIKK366 pKa = 7.01FTKK369 pKa = 7.93PTIVIQSVPTSVSQILLVADD389 pKa = 4.93MIRR392 pKa = 11.84TITPEE397 pKa = 3.86LVSEE401 pKa = 4.19YY402 pKa = 10.3TSAIKK407 pKa = 10.02PIIEE411 pKa = 4.26MIAYY415 pKa = 9.71SVVTEE420 pKa = 3.99VLKK423 pKa = 11.02SVSVSASEE431 pKa = 4.14EE432 pKa = 3.98FLISTVYY439 pKa = 10.52EE440 pKa = 4.49VVQKK444 pKa = 11.05ALVPIYY450 pKa = 10.01TSASEE455 pKa = 4.2EE456 pKa = 3.93FLNTVIYY463 pKa = 7.48EE464 pKa = 4.35TVKK467 pKa = 10.79EE468 pKa = 4.38VVTSLKK474 pKa = 8.05TTQSEE479 pKa = 4.17ITMVSYY485 pKa = 10.74VQEE488 pKa = 4.38KK489 pKa = 8.1VTPTQFSISSSQSEE503 pKa = 4.5TTMVSYY509 pKa = 10.55VQEE512 pKa = 4.92KK513 pKa = 8.48ITPTQLPVSSSQSEE527 pKa = 4.55GIMVSSVYY535 pKa = 9.11QQITAVVVPEE545 pKa = 4.41TTTSPAEE552 pKa = 4.2TIVNAIISCPFVDD565 pKa = 5.91AKK567 pKa = 10.13NQCYY571 pKa = 6.33TTGYY575 pKa = 9.1TIYY578 pKa = 10.71LFNNTSTTIPSGYY591 pKa = 9.21PIPIWIYY598 pKa = 10.1VDD600 pKa = 3.87GANSTYY606 pKa = 11.15SNVRR610 pKa = 11.84LSYY613 pKa = 10.83NGNEE617 pKa = 4.03LYY619 pKa = 9.75TWVEE623 pKa = 4.2EE624 pKa = 4.27IVGGSALMWVVLPTDD639 pKa = 4.41LSTVIQIDD647 pKa = 3.51VWVGDD652 pKa = 3.51SSFVYY657 pKa = 10.49DD658 pKa = 4.58GVAGAYY664 pKa = 9.51KK665 pKa = 10.37EE666 pKa = 4.17IAGCSNDD673 pKa = 3.44NGSKK677 pKa = 10.23IFTLYY682 pKa = 11.28DD683 pKa = 3.64NFCGTSLNTDD693 pKa = 2.36IWNLYY698 pKa = 9.26VSGDD702 pKa = 3.38YY703 pKa = 10.73SYY705 pKa = 11.52EE706 pKa = 4.06VNNDD710 pKa = 3.95LIINVSQGQTGYY722 pKa = 11.13VVLQSKK728 pKa = 7.27QTFGEE733 pKa = 4.85GYY735 pKa = 8.49MFEE738 pKa = 4.49VMSSTSNFITNIRR751 pKa = 11.84TAQPSISVGNGAEE764 pKa = 4.05YY765 pKa = 11.0AFTDD769 pKa = 5.3GIISSSSPQEE779 pKa = 3.78TADD782 pKa = 3.7FSINGNCVPYY792 pKa = 10.59YY793 pKa = 10.15VVEE796 pKa = 4.86LGTQSLTCPSSCPSSSSSFCEE817 pKa = 3.73TSYY820 pKa = 10.45ATYY823 pKa = 10.86AVIGINYY830 pKa = 7.75PFRR833 pKa = 11.84KK834 pKa = 9.88LISGLFYY841 pKa = 10.93QNNNVALVVDD851 pKa = 4.43RR852 pKa = 11.84VMLVNSLPSQSISGNVYY869 pKa = 8.92MYY871 pKa = 11.07LGTAYY876 pKa = 10.86SDD878 pKa = 3.26ATGNLTLTYY887 pKa = 9.84HH888 pKa = 6.79FARR891 pKa = 11.84VYY893 pKa = 10.99PMIAPPFPASQSGYY907 pKa = 9.94IVDD910 pKa = 4.86SYY912 pKa = 11.54WEE914 pKa = 4.23FNGSSSYY921 pKa = 10.23MVVEE925 pKa = 4.7LPQVSSSSHH934 pKa = 4.63TFNFTVTYY942 pKa = 10.04AVWFRR947 pKa = 11.84TTSGTGMILSDD958 pKa = 3.84NQSASSPCYY967 pKa = 10.14DD968 pKa = 3.95YY969 pKa = 11.96YY970 pKa = 11.83NDD972 pKa = 3.88ILSIVNGKK980 pKa = 8.97LVAGYY985 pKa = 9.27CCNGTFVTTSDD996 pKa = 3.92TYY998 pKa = 11.94NDD1000 pKa = 4.47GNWHH1004 pKa = 5.89FAVVILDD1011 pKa = 3.71PSNNSITLYY1020 pKa = 10.97VDD1022 pKa = 3.19GQEE1025 pKa = 4.17IGSATVSSSLEE1036 pKa = 3.95PPNPYY1041 pKa = 10.09YY1042 pKa = 10.93LWIGYY1047 pKa = 9.26GSSCAGWADD1056 pKa = 3.79EE1057 pKa = 5.27GISGYY1062 pKa = 10.9FNGDD1066 pKa = 3.47IAWVAVFNTALSSSQIQEE1084 pKa = 4.0IYY1086 pKa = 10.99QMGPYY1091 pKa = 10.22SIDD1094 pKa = 3.23PLLFTDD1100 pKa = 5.8NIVGWWTPWNGPPDD1114 pKa = 4.64LSTADD1119 pKa = 4.98DD1120 pKa = 3.68SLTYY1124 pKa = 10.69YY1125 pKa = 10.79NISTPSFVGYY1135 pKa = 10.21VGVYY1139 pKa = 8.42PAIDD1143 pKa = 3.48AFYY1146 pKa = 10.86SDD1148 pKa = 3.77ARR1150 pKa = 11.84TTFVLYY1156 pKa = 8.7NTYY1159 pKa = 10.69SSLMVTLTSPTTGQYY1174 pKa = 10.77GGVIFSYY1181 pKa = 10.5SYY1183 pKa = 8.8TQGNVFTVSGLFTLDD1198 pKa = 3.9NPSSCPADD1206 pKa = 3.22GFAFAIFSSGFDD1218 pKa = 3.14TSNIVVPSISGCKK1231 pKa = 9.01FQGSMSIPEE1240 pKa = 4.26TSSPTILVQYY1250 pKa = 9.89DD1251 pKa = 3.99PYY1253 pKa = 10.31WAEE1256 pKa = 3.91PCAGNQVGTGVNISVVNGTSVTEE1279 pKa = 4.12IVCGAGSGTFNTNVQSGNEE1298 pKa = 3.68MFYY1301 pKa = 9.97IVSYY1305 pKa = 10.25IPSSNEE1311 pKa = 3.02VCYY1314 pKa = 10.6TMFFIPNNQFVTGCTSLGSDD1334 pKa = 4.15FSAPSSGTYY1343 pKa = 9.87YY1344 pKa = 10.72LAVAGATGGQYY1355 pKa = 10.74AQWLFDD1361 pKa = 3.42ARR1363 pKa = 11.84TLHH1366 pKa = 7.08ISQQ1369 pKa = 4.07
MM1 pKa = 7.68SCGEE5 pKa = 4.8LPNASCTPTPNWEE18 pKa = 4.25CSDD21 pKa = 3.8VEE23 pKa = 5.31CFQQCINQPTFEE35 pKa = 4.63QFYY38 pKa = 9.5EE39 pKa = 4.38CCIQCALAHH48 pKa = 6.14YY49 pKa = 9.62IAEE52 pKa = 4.68AKK54 pKa = 10.57DD55 pKa = 3.31HH56 pKa = 5.05VTILVANAVSMIGTEE71 pKa = 4.18YY72 pKa = 10.6IPVFDD77 pKa = 3.95QMQMFPAHH85 pKa = 7.15LEE87 pKa = 3.98TLQGKK92 pKa = 7.47EE93 pKa = 3.77QVTVRR98 pKa = 11.84DD99 pKa = 3.4LVFPQNYY106 pKa = 8.08PVKK109 pKa = 10.5KK110 pKa = 7.46YY111 pKa = 8.92TYY113 pKa = 9.33YY114 pKa = 11.18EE115 pKa = 3.97NVQVNDD121 pKa = 3.06IGQPCSSPTGFCVPPGVTASEE142 pKa = 4.17EE143 pKa = 3.89LWFVNEE149 pKa = 4.03GSVVIIPSTLPPTTVSTFSYY169 pKa = 8.58ITPPPPISFQVTYY182 pKa = 7.7PTLVVPPPPPSPPPTTNILVVSKK205 pKa = 9.54PCPEE209 pKa = 3.95VCPGCICYY217 pKa = 10.68YY218 pKa = 9.68MLGIDD223 pKa = 4.76IVDD226 pKa = 3.85PTDD229 pKa = 3.12AVYY232 pKa = 10.61NYY234 pKa = 9.99QEE236 pKa = 4.3TPTGVVQQVPQNEE249 pKa = 4.36YY250 pKa = 10.98VLPSDD255 pKa = 4.12LLSIEE260 pKa = 4.42EE261 pKa = 4.58APSHH265 pKa = 4.59TTEE268 pKa = 4.11TEE270 pKa = 4.11TVEE273 pKa = 4.0EE274 pKa = 4.65SISFTLYY281 pKa = 10.59YY282 pKa = 10.73VVTTTLPVLDD292 pKa = 4.17VNFVYY297 pKa = 10.03PVTLIRR303 pKa = 11.84EE304 pKa = 4.47VEE306 pKa = 4.32SVTASRR312 pKa = 11.84SIQTVYY318 pKa = 10.48FSYY321 pKa = 10.35PVSKK325 pKa = 10.43QVFSEE330 pKa = 4.45YY331 pKa = 8.61VTASSISVSAYY342 pKa = 7.48FQPIVKK348 pKa = 9.84LVQEE352 pKa = 4.47SEE354 pKa = 4.6VTVSSIPILQIKK366 pKa = 7.01FTKK369 pKa = 7.93PTIVIQSVPTSVSQILLVADD389 pKa = 4.93MIRR392 pKa = 11.84TITPEE397 pKa = 3.86LVSEE401 pKa = 4.19YY402 pKa = 10.3TSAIKK407 pKa = 10.02PIIEE411 pKa = 4.26MIAYY415 pKa = 9.71SVVTEE420 pKa = 3.99VLKK423 pKa = 11.02SVSVSASEE431 pKa = 4.14EE432 pKa = 3.98FLISTVYY439 pKa = 10.52EE440 pKa = 4.49VVQKK444 pKa = 11.05ALVPIYY450 pKa = 10.01TSASEE455 pKa = 4.2EE456 pKa = 3.93FLNTVIYY463 pKa = 7.48EE464 pKa = 4.35TVKK467 pKa = 10.79EE468 pKa = 4.38VVTSLKK474 pKa = 8.05TTQSEE479 pKa = 4.17ITMVSYY485 pKa = 10.74VQEE488 pKa = 4.38KK489 pKa = 8.1VTPTQFSISSSQSEE503 pKa = 4.5TTMVSYY509 pKa = 10.55VQEE512 pKa = 4.92KK513 pKa = 8.48ITPTQLPVSSSQSEE527 pKa = 4.55GIMVSSVYY535 pKa = 9.11QQITAVVVPEE545 pKa = 4.41TTTSPAEE552 pKa = 4.2TIVNAIISCPFVDD565 pKa = 5.91AKK567 pKa = 10.13NQCYY571 pKa = 6.33TTGYY575 pKa = 9.1TIYY578 pKa = 10.71LFNNTSTTIPSGYY591 pKa = 9.21PIPIWIYY598 pKa = 10.1VDD600 pKa = 3.87GANSTYY606 pKa = 11.15SNVRR610 pKa = 11.84LSYY613 pKa = 10.83NGNEE617 pKa = 4.03LYY619 pKa = 9.75TWVEE623 pKa = 4.2EE624 pKa = 4.27IVGGSALMWVVLPTDD639 pKa = 4.41LSTVIQIDD647 pKa = 3.51VWVGDD652 pKa = 3.51SSFVYY657 pKa = 10.49DD658 pKa = 4.58GVAGAYY664 pKa = 9.51KK665 pKa = 10.37EE666 pKa = 4.17IAGCSNDD673 pKa = 3.44NGSKK677 pKa = 10.23IFTLYY682 pKa = 11.28DD683 pKa = 3.64NFCGTSLNTDD693 pKa = 2.36IWNLYY698 pKa = 9.26VSGDD702 pKa = 3.38YY703 pKa = 10.73SYY705 pKa = 11.52EE706 pKa = 4.06VNNDD710 pKa = 3.95LIINVSQGQTGYY722 pKa = 11.13VVLQSKK728 pKa = 7.27QTFGEE733 pKa = 4.85GYY735 pKa = 8.49MFEE738 pKa = 4.49VMSSTSNFITNIRR751 pKa = 11.84TAQPSISVGNGAEE764 pKa = 4.05YY765 pKa = 11.0AFTDD769 pKa = 5.3GIISSSSPQEE779 pKa = 3.78TADD782 pKa = 3.7FSINGNCVPYY792 pKa = 10.59YY793 pKa = 10.15VVEE796 pKa = 4.86LGTQSLTCPSSCPSSSSSFCEE817 pKa = 3.73TSYY820 pKa = 10.45ATYY823 pKa = 10.86AVIGINYY830 pKa = 7.75PFRR833 pKa = 11.84KK834 pKa = 9.88LISGLFYY841 pKa = 10.93QNNNVALVVDD851 pKa = 4.43RR852 pKa = 11.84VMLVNSLPSQSISGNVYY869 pKa = 8.92MYY871 pKa = 11.07LGTAYY876 pKa = 10.86SDD878 pKa = 3.26ATGNLTLTYY887 pKa = 9.84HH888 pKa = 6.79FARR891 pKa = 11.84VYY893 pKa = 10.99PMIAPPFPASQSGYY907 pKa = 9.94IVDD910 pKa = 4.86SYY912 pKa = 11.54WEE914 pKa = 4.23FNGSSSYY921 pKa = 10.23MVVEE925 pKa = 4.7LPQVSSSSHH934 pKa = 4.63TFNFTVTYY942 pKa = 10.04AVWFRR947 pKa = 11.84TTSGTGMILSDD958 pKa = 3.84NQSASSPCYY967 pKa = 10.14DD968 pKa = 3.95YY969 pKa = 11.96YY970 pKa = 11.83NDD972 pKa = 3.88ILSIVNGKK980 pKa = 8.97LVAGYY985 pKa = 9.27CCNGTFVTTSDD996 pKa = 3.92TYY998 pKa = 11.94NDD1000 pKa = 4.47GNWHH1004 pKa = 5.89FAVVILDD1011 pKa = 3.71PSNNSITLYY1020 pKa = 10.97VDD1022 pKa = 3.19GQEE1025 pKa = 4.17IGSATVSSSLEE1036 pKa = 3.95PPNPYY1041 pKa = 10.09YY1042 pKa = 10.93LWIGYY1047 pKa = 9.26GSSCAGWADD1056 pKa = 3.79EE1057 pKa = 5.27GISGYY1062 pKa = 10.9FNGDD1066 pKa = 3.47IAWVAVFNTALSSSQIQEE1084 pKa = 4.0IYY1086 pKa = 10.99QMGPYY1091 pKa = 10.22SIDD1094 pKa = 3.23PLLFTDD1100 pKa = 5.8NIVGWWTPWNGPPDD1114 pKa = 4.64LSTADD1119 pKa = 4.98DD1120 pKa = 3.68SLTYY1124 pKa = 10.69YY1125 pKa = 10.79NISTPSFVGYY1135 pKa = 10.21VGVYY1139 pKa = 8.42PAIDD1143 pKa = 3.48AFYY1146 pKa = 10.86SDD1148 pKa = 3.77ARR1150 pKa = 11.84TTFVLYY1156 pKa = 8.7NTYY1159 pKa = 10.69SSLMVTLTSPTTGQYY1174 pKa = 10.77GGVIFSYY1181 pKa = 10.5SYY1183 pKa = 8.8TQGNVFTVSGLFTLDD1198 pKa = 3.9NPSSCPADD1206 pKa = 3.22GFAFAIFSSGFDD1218 pKa = 3.14TSNIVVPSISGCKK1231 pKa = 9.01FQGSMSIPEE1240 pKa = 4.26TSSPTILVQYY1250 pKa = 9.89DD1251 pKa = 3.99PYY1253 pKa = 10.31WAEE1256 pKa = 3.91PCAGNQVGTGVNISVVNGTSVTEE1279 pKa = 4.12IVCGAGSGTFNTNVQSGNEE1298 pKa = 3.68MFYY1301 pKa = 9.97IVSYY1305 pKa = 10.25IPSSNEE1311 pKa = 3.02VCYY1314 pKa = 10.6TMFFIPNNQFVTGCTSLGSDD1334 pKa = 4.15FSAPSSGTYY1343 pKa = 9.87YY1344 pKa = 10.72LAVAGATGGQYY1355 pKa = 10.74AQWLFDD1361 pKa = 3.42ARR1363 pKa = 11.84TLHH1366 pKa = 7.08ISQQ1369 pKa = 4.07
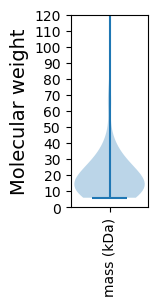
Molecular weight: 149.31 kDa
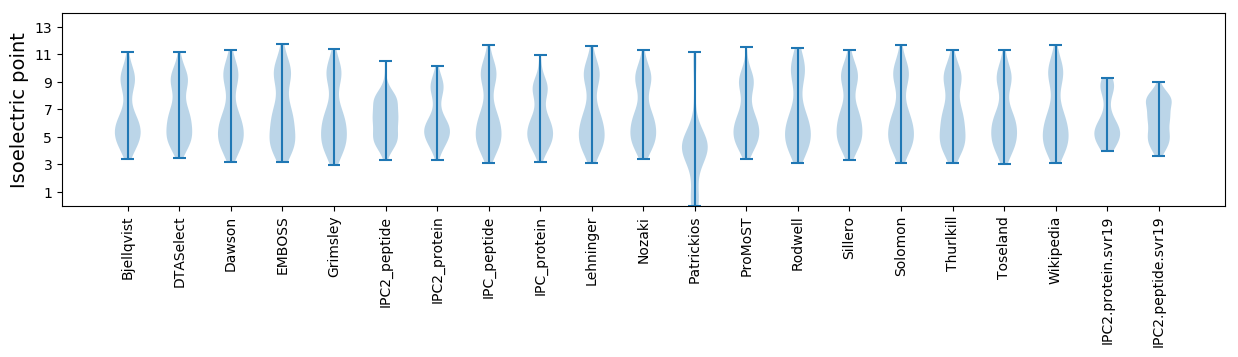
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7WKJ2|A7WKJ2_9VIRU Uncharacterized protein OS=Acidianus filamentous virus 6 OX=346882 PE=4 SV=1
MM1 pKa = 7.16MKK3 pKa = 8.65ITKK6 pKa = 6.76TTFVRR11 pKa = 11.84LCFWALSQARR21 pKa = 11.84MSGIKK26 pKa = 9.55VVPRR30 pKa = 11.84IMLRR34 pKa = 11.84FLDD37 pKa = 4.06EE38 pKa = 4.33VKK40 pKa = 10.66ASPRR44 pKa = 11.84TKK46 pKa = 10.47EE47 pKa = 3.91RR48 pKa = 11.84LLLLIDD54 pKa = 4.24EE55 pKa = 5.49LDD57 pKa = 3.53TGKK60 pKa = 8.47ITPRR64 pKa = 11.84EE65 pKa = 3.71FARR68 pKa = 11.84RR69 pKa = 11.84LFLEE73 pKa = 4.31YY74 pKa = 9.92PRR76 pKa = 11.84PTRR79 pKa = 11.84KK80 pKa = 9.94GRR82 pKa = 11.84VLSLKK87 pKa = 8.45EE88 pKa = 4.02FKK90 pKa = 10.57RR91 pKa = 11.84ITSLGKK97 pKa = 10.09
MM1 pKa = 7.16MKK3 pKa = 8.65ITKK6 pKa = 6.76TTFVRR11 pKa = 11.84LCFWALSQARR21 pKa = 11.84MSGIKK26 pKa = 9.55VVPRR30 pKa = 11.84IMLRR34 pKa = 11.84FLDD37 pKa = 4.06EE38 pKa = 4.33VKK40 pKa = 10.66ASPRR44 pKa = 11.84TKK46 pKa = 10.47EE47 pKa = 3.91RR48 pKa = 11.84LLLLIDD54 pKa = 4.24EE55 pKa = 5.49LDD57 pKa = 3.53TGKK60 pKa = 8.47ITPRR64 pKa = 11.84EE65 pKa = 3.71FARR68 pKa = 11.84RR69 pKa = 11.84LFLEE73 pKa = 4.31YY74 pKa = 9.92PRR76 pKa = 11.84PTRR79 pKa = 11.84KK80 pKa = 9.94GRR82 pKa = 11.84VLSLKK87 pKa = 8.45EE88 pKa = 4.02FKK90 pKa = 10.57RR91 pKa = 11.84ITSLGKK97 pKa = 10.09
Molecular weight: 11.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11980 |
49 |
1369 |
181.5 |
20.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.925 ± 0.238 | 1.377 ± 0.167 |
4.624 ± 0.232 | 7.429 ± 0.451 |
4.416 ± 0.173 | 5.242 ± 0.22 |
1.311 ± 0.162 | 8.865 ± 0.4 |
7.321 ± 0.724 | 8.664 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.158 | 5.192 ± 0.223 |
3.973 ± 0.355 | 3.18 ± 0.243 |
3.681 ± 0.393 | 6.619 ± 0.722 |
5.985 ± 0.461 | 7.98 ± 0.427 |
1.018 ± 0.104 | 5.735 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
