
Thalassotalea agarivorans (Thalassomonas agarivorans)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Colwelliaceae; Thalassotalea
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
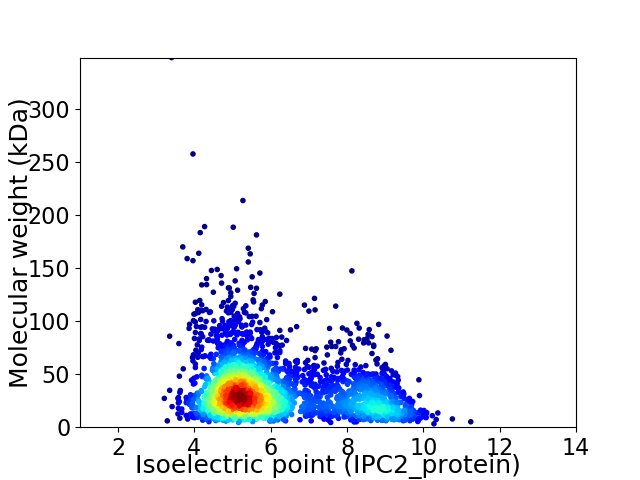
Virtual 2D-PAGE plot for 2994 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0BKS3|A0A1I0BKS3_THASX Hydrolase_4 domain-containing protein OS=Thalassotalea agarivorans OX=349064 GN=SAMN05660429_00982 PE=4 SV=1
MM1 pKa = 6.92ITSSKK6 pKa = 10.42KK7 pKa = 9.63IVSAMLSTSLWIGVASTAFAEE28 pKa = 4.61TTNVEE33 pKa = 3.97AEE35 pKa = 4.64GYY37 pKa = 7.94STIGGTYY44 pKa = 9.73QDD46 pKa = 5.21GNPQPINIYY55 pKa = 10.0SVNGVQAINFVNRR68 pKa = 11.84GDD70 pKa = 3.52FAEE73 pKa = 4.29YY74 pKa = 10.33DD75 pKa = 3.88VSVSTAGEE83 pKa = 3.76YY84 pKa = 10.53SIEE87 pKa = 3.91YY88 pKa = 10.52LIGTSIASGSAIEE101 pKa = 4.53ISVLVDD107 pKa = 3.95GNWQSAGSTNVPLGHH122 pKa = 7.29WDD124 pKa = 3.42NFQALAATNNISLAQGTNRR143 pKa = 11.84IKK145 pKa = 9.67ITGAGTHH152 pKa = 6.15DD153 pKa = 3.73WQWNLDD159 pKa = 3.76AFSLTLEE166 pKa = 4.4TPEE169 pKa = 5.56NPDD172 pKa = 4.35NPDD175 pKa = 3.51NPDD178 pKa = 3.52NPDD181 pKa = 3.44NPDD184 pKa = 3.49NPDD187 pKa = 4.3DD188 pKa = 4.94GNTGQPGTPFTIEE201 pKa = 3.72MEE203 pKa = 4.35AFNATGSDD211 pKa = 3.68DD212 pKa = 3.59PRR214 pKa = 11.84AQGMIIGEE222 pKa = 3.96RR223 pKa = 11.84GYY225 pKa = 10.03PEE227 pKa = 5.41DD228 pKa = 3.22KK229 pKa = 10.14HH230 pKa = 6.02TVVDD234 pKa = 4.51SNQTTDD240 pKa = 2.77WVDD243 pKa = 3.27YY244 pKa = 10.72NINFPVSGNYY254 pKa = 9.55RR255 pKa = 11.84IEE257 pKa = 4.08MLASGQTSHH266 pKa = 6.08ATAILFVDD274 pKa = 3.81NVQINEE280 pKa = 4.21VPVDD284 pKa = 3.53TGNQAVFQDD293 pKa = 4.62FEE295 pKa = 4.75LTDD298 pKa = 3.35STYY301 pKa = 10.89ISAGAHH307 pKa = 5.84TIRR310 pKa = 11.84VQSGSQINEE319 pKa = 4.07FSWMWFGDD327 pKa = 3.49ALTFTPLDD335 pKa = 4.03GGSTDD340 pKa = 4.44GDD342 pKa = 3.74ADD344 pKa = 3.74NDD346 pKa = 4.19GVLDD350 pKa = 5.16SVDD353 pKa = 3.52TCPNTPAGAQVDD365 pKa = 4.07ANGCEE370 pKa = 4.04IIVDD374 pKa = 3.83NDD376 pKa = 3.34TDD378 pKa = 3.92NDD380 pKa = 4.2GVDD383 pKa = 3.88NSIDD387 pKa = 3.52QCPNTPAGAQVDD399 pKa = 4.09ANGCEE404 pKa = 3.8IVAVVDD410 pKa = 3.72ADD412 pKa = 3.65NDD414 pKa = 4.12GVEE417 pKa = 5.21DD418 pKa = 4.5SLDD421 pKa = 3.53MCPNTPAGAPVNGQGCADD439 pKa = 3.85SQLDD443 pKa = 3.57ADD445 pKa = 4.36NDD447 pKa = 4.3GVSDD451 pKa = 6.62DD452 pKa = 4.87IDD454 pKa = 3.62QCPSTPAGSVVDD466 pKa = 3.91GTGCIVVTPPADD478 pKa = 3.43SDD480 pKa = 3.79NDD482 pKa = 4.0GVADD486 pKa = 4.21TLDD489 pKa = 3.73MCPNTATGLTVDD501 pKa = 3.9SQGCALSQLDD511 pKa = 3.97SDD513 pKa = 4.39NDD515 pKa = 3.93GVTDD519 pKa = 6.85DD520 pKa = 5.44IDD522 pKa = 3.48QCANTPSGEE531 pKa = 4.19TANATGCSSSQEE543 pKa = 4.21GGGTDD548 pKa = 3.63PGTPQPGLLYY558 pKa = 11.0GEE560 pKa = 5.06LAGAMNVSDD569 pKa = 4.71TNPNWEE575 pKa = 4.16RR576 pKa = 11.84TTDD579 pKa = 3.84LLQTEE584 pKa = 4.91DD585 pKa = 4.07SIKK588 pKa = 11.01GNTTEE593 pKa = 4.2VYY595 pKa = 9.97TGFIYY600 pKa = 10.85DD601 pKa = 3.74ADD603 pKa = 3.87GHH605 pKa = 6.51ISFYY609 pKa = 10.14EE610 pKa = 4.46HH611 pKa = 6.94IDD613 pKa = 3.45DD614 pKa = 5.24SVRR617 pKa = 11.84LYY619 pKa = 10.53IDD621 pKa = 3.54GVLVLSNDD629 pKa = 2.74SWEE632 pKa = 4.61ASSQTTDD639 pKa = 3.62LNLTPGTHH647 pKa = 6.53EE648 pKa = 3.7IEE650 pKa = 4.19LRR652 pKa = 11.84IGNADD657 pKa = 3.67GGSGAVDD664 pKa = 4.42GIGFGIDD671 pKa = 2.98VDD673 pKa = 3.83GGTNFVHH680 pKa = 7.4PSTLSEE686 pKa = 4.58SIFTSVGEE694 pKa = 4.19EE695 pKa = 3.89TGNPDD700 pKa = 4.41LEE702 pKa = 4.37QEE704 pKa = 3.85GDD706 pKa = 3.6IIVEE710 pKa = 4.05LEE712 pKa = 4.17SFVFTSTNGRR722 pKa = 11.84VGGDD726 pKa = 3.35SVEE729 pKa = 4.47GFSPTATGVNWVTNGDD745 pKa = 3.69YY746 pKa = 11.08GDD748 pKa = 3.86YY749 pKa = 9.75MVTFEE754 pKa = 5.2EE755 pKa = 4.89PGTYY759 pKa = 9.38SAYY762 pKa = 8.88ITISAANDD770 pKa = 3.45GSYY773 pKa = 10.64GARR776 pKa = 11.84VDD778 pKa = 3.47VDD780 pKa = 3.51GWPVAWGYY788 pKa = 11.05FGGTGSWDD796 pKa = 3.32VSSEE800 pKa = 3.57NLLYY804 pKa = 10.94GGTFVVEE811 pKa = 3.88QAGEE815 pKa = 3.97KK816 pKa = 9.34VVRR819 pKa = 11.84VEE821 pKa = 5.28AIGGSDD827 pKa = 3.53WQWSGDD833 pKa = 3.16RR834 pKa = 11.84VRR836 pKa = 11.84FTRR839 pKa = 11.84LGDD842 pKa = 3.31VTAIPSPIYY851 pKa = 10.84NPDD854 pKa = 3.21DD855 pKa = 3.96HH856 pKa = 7.09FVAEE860 pKa = 4.25IQGPQTDD867 pKa = 3.52VTYY870 pKa = 10.65LKK872 pKa = 10.73KK873 pKa = 10.31PVEE876 pKa = 3.9IPANKK881 pKa = 9.53KK882 pKa = 8.2VLKK885 pKa = 10.35SDD887 pKa = 2.47VWYY890 pKa = 8.63TYY892 pKa = 8.87PQNRR896 pKa = 11.84EE897 pKa = 3.72LEE899 pKa = 4.62GYY901 pKa = 10.75DD902 pKa = 3.25NFGATGAFWGHH913 pKa = 6.31PPEE916 pKa = 6.01HH917 pKa = 7.32DD918 pKa = 4.04FYY920 pKa = 11.46DD921 pKa = 3.9DD922 pKa = 3.6TVIMDD927 pKa = 4.26WAVDD931 pKa = 3.43AVYY934 pKa = 10.73AFQAEE939 pKa = 5.03GYY941 pKa = 8.67EE942 pKa = 4.05YY943 pKa = 9.77TARR946 pKa = 11.84GEE948 pKa = 4.11FDD950 pKa = 2.68WGYY953 pKa = 11.45GWFTEE958 pKa = 4.56YY959 pKa = 7.85TTNPQPHH966 pKa = 5.68YY967 pKa = 11.23VRR969 pKa = 11.84TLDD972 pKa = 3.67DD973 pKa = 3.98RR974 pKa = 11.84NVRR977 pKa = 11.84MTFMGYY983 pKa = 10.5LSHH986 pKa = 7.94DD987 pKa = 3.87GYY989 pKa = 11.54NNNWLSNHH997 pKa = 5.71SPAFVPFMKK1006 pKa = 10.46SQVDD1010 pKa = 3.78QILKK1014 pKa = 10.51ANPDD1018 pKa = 3.23KK1019 pKa = 11.52LMFDD1023 pKa = 3.57TQTNSTRR1030 pKa = 11.84STDD1033 pKa = 3.25MRR1035 pKa = 11.84DD1036 pKa = 3.24FGGDD1040 pKa = 3.54FSPYY1044 pKa = 9.96AMEE1047 pKa = 4.92NFRR1050 pKa = 11.84VWLSKK1055 pKa = 10.49KK1056 pKa = 10.39YY1057 pKa = 9.44STGEE1061 pKa = 3.73LAALGINDD1069 pKa = 4.36INSFDD1074 pKa = 3.68YY1075 pKa = 11.46GDD1077 pKa = 3.75FLRR1080 pKa = 11.84AQGVTHH1086 pKa = 6.5TSWSNAGDD1094 pKa = 3.76TLSGNIPLQEE1104 pKa = 4.06DD1105 pKa = 4.21YY1106 pKa = 10.91IYY1108 pKa = 10.85FNRR1111 pKa = 11.84DD1112 pKa = 1.96VWNQKK1117 pKa = 8.47FAEE1120 pKa = 4.14VLDD1123 pKa = 4.67YY1124 pKa = 10.77IRR1126 pKa = 11.84QQQPDD1131 pKa = 3.68IEE1133 pKa = 5.14IGASTHH1139 pKa = 5.75LFEE1142 pKa = 5.23SRR1144 pKa = 11.84GYY1146 pKa = 10.33VFNEE1150 pKa = 3.35NLTFLSGEE1158 pKa = 4.24LNLGARR1164 pKa = 11.84TTISEE1169 pKa = 4.63LPTNILVHH1177 pKa = 6.42LKK1179 pKa = 10.2GAQAVDD1185 pKa = 3.12KK1186 pKa = 9.61TLVYY1190 pKa = 10.12FPYY1193 pKa = 9.78PWEE1196 pKa = 4.08FDD1198 pKa = 3.43EE1199 pKa = 6.74LRR1201 pKa = 11.84LQDD1204 pKa = 3.35APRR1207 pKa = 11.84FGRR1210 pKa = 11.84GWVAQAYY1217 pKa = 9.84AYY1219 pKa = 10.24GGLFSIPANVWVGGEE1234 pKa = 3.93VWTWSPGADD1243 pKa = 3.33NYY1245 pKa = 11.19RR1246 pKa = 11.84DD1247 pKa = 2.94IYY1249 pKa = 11.15LFVRR1253 pKa = 11.84AQADD1257 pKa = 3.7LLDD1260 pKa = 5.57DD1261 pKa = 3.66YY1262 pKa = 11.46TSYY1265 pKa = 11.69SKK1267 pKa = 11.15VGLVHH1272 pKa = 7.3AMYY1275 pKa = 11.07SSMKK1279 pKa = 10.49AGFIDD1284 pKa = 4.46GGNQIQSSTKK1294 pKa = 10.39LLTEE1298 pKa = 4.09GNINFDD1304 pKa = 3.9LLVFGDD1310 pKa = 3.51EE1311 pKa = 4.94GYY1313 pKa = 9.94PVVPRR1318 pKa = 11.84PEE1320 pKa = 5.09DD1321 pKa = 3.49FDD1323 pKa = 4.58KK1324 pKa = 10.84FDD1326 pKa = 5.44HH1327 pKa = 6.72IFFDD1331 pKa = 4.82GDD1333 pKa = 3.54QQYY1336 pKa = 7.85LTAEE1340 pKa = 4.11QQALLDD1346 pKa = 3.79QQGDD1350 pKa = 3.81KK1351 pKa = 10.67VRR1353 pKa = 11.84HH1354 pKa = 5.17IGQRR1358 pKa = 11.84GTVSGIEE1365 pKa = 3.65ITVSINGTEE1374 pKa = 4.17SNEE1377 pKa = 4.15TVSAVSRR1384 pKa = 11.84IHH1386 pKa = 5.76EE1387 pKa = 4.15TDD1389 pKa = 2.75AAAPYY1394 pKa = 9.81VVHH1397 pKa = 6.67LVNRR1401 pKa = 11.84PFAGGVTPTLNNVEE1415 pKa = 4.2VAIPQSYY1422 pKa = 9.36FPEE1425 pKa = 4.52VVTGATLHH1433 pKa = 6.63LPDD1436 pKa = 4.49GTSTPLTLSTNADD1449 pKa = 3.39GDD1451 pKa = 4.0VVLPVNNLEE1460 pKa = 3.82VWGILEE1466 pKa = 4.3LAHH1469 pKa = 7.01
MM1 pKa = 6.92ITSSKK6 pKa = 10.42KK7 pKa = 9.63IVSAMLSTSLWIGVASTAFAEE28 pKa = 4.61TTNVEE33 pKa = 3.97AEE35 pKa = 4.64GYY37 pKa = 7.94STIGGTYY44 pKa = 9.73QDD46 pKa = 5.21GNPQPINIYY55 pKa = 10.0SVNGVQAINFVNRR68 pKa = 11.84GDD70 pKa = 3.52FAEE73 pKa = 4.29YY74 pKa = 10.33DD75 pKa = 3.88VSVSTAGEE83 pKa = 3.76YY84 pKa = 10.53SIEE87 pKa = 3.91YY88 pKa = 10.52LIGTSIASGSAIEE101 pKa = 4.53ISVLVDD107 pKa = 3.95GNWQSAGSTNVPLGHH122 pKa = 7.29WDD124 pKa = 3.42NFQALAATNNISLAQGTNRR143 pKa = 11.84IKK145 pKa = 9.67ITGAGTHH152 pKa = 6.15DD153 pKa = 3.73WQWNLDD159 pKa = 3.76AFSLTLEE166 pKa = 4.4TPEE169 pKa = 5.56NPDD172 pKa = 4.35NPDD175 pKa = 3.51NPDD178 pKa = 3.52NPDD181 pKa = 3.44NPDD184 pKa = 3.49NPDD187 pKa = 4.3DD188 pKa = 4.94GNTGQPGTPFTIEE201 pKa = 3.72MEE203 pKa = 4.35AFNATGSDD211 pKa = 3.68DD212 pKa = 3.59PRR214 pKa = 11.84AQGMIIGEE222 pKa = 3.96RR223 pKa = 11.84GYY225 pKa = 10.03PEE227 pKa = 5.41DD228 pKa = 3.22KK229 pKa = 10.14HH230 pKa = 6.02TVVDD234 pKa = 4.51SNQTTDD240 pKa = 2.77WVDD243 pKa = 3.27YY244 pKa = 10.72NINFPVSGNYY254 pKa = 9.55RR255 pKa = 11.84IEE257 pKa = 4.08MLASGQTSHH266 pKa = 6.08ATAILFVDD274 pKa = 3.81NVQINEE280 pKa = 4.21VPVDD284 pKa = 3.53TGNQAVFQDD293 pKa = 4.62FEE295 pKa = 4.75LTDD298 pKa = 3.35STYY301 pKa = 10.89ISAGAHH307 pKa = 5.84TIRR310 pKa = 11.84VQSGSQINEE319 pKa = 4.07FSWMWFGDD327 pKa = 3.49ALTFTPLDD335 pKa = 4.03GGSTDD340 pKa = 4.44GDD342 pKa = 3.74ADD344 pKa = 3.74NDD346 pKa = 4.19GVLDD350 pKa = 5.16SVDD353 pKa = 3.52TCPNTPAGAQVDD365 pKa = 4.07ANGCEE370 pKa = 4.04IIVDD374 pKa = 3.83NDD376 pKa = 3.34TDD378 pKa = 3.92NDD380 pKa = 4.2GVDD383 pKa = 3.88NSIDD387 pKa = 3.52QCPNTPAGAQVDD399 pKa = 4.09ANGCEE404 pKa = 3.8IVAVVDD410 pKa = 3.72ADD412 pKa = 3.65NDD414 pKa = 4.12GVEE417 pKa = 5.21DD418 pKa = 4.5SLDD421 pKa = 3.53MCPNTPAGAPVNGQGCADD439 pKa = 3.85SQLDD443 pKa = 3.57ADD445 pKa = 4.36NDD447 pKa = 4.3GVSDD451 pKa = 6.62DD452 pKa = 4.87IDD454 pKa = 3.62QCPSTPAGSVVDD466 pKa = 3.91GTGCIVVTPPADD478 pKa = 3.43SDD480 pKa = 3.79NDD482 pKa = 4.0GVADD486 pKa = 4.21TLDD489 pKa = 3.73MCPNTATGLTVDD501 pKa = 3.9SQGCALSQLDD511 pKa = 3.97SDD513 pKa = 4.39NDD515 pKa = 3.93GVTDD519 pKa = 6.85DD520 pKa = 5.44IDD522 pKa = 3.48QCANTPSGEE531 pKa = 4.19TANATGCSSSQEE543 pKa = 4.21GGGTDD548 pKa = 3.63PGTPQPGLLYY558 pKa = 11.0GEE560 pKa = 5.06LAGAMNVSDD569 pKa = 4.71TNPNWEE575 pKa = 4.16RR576 pKa = 11.84TTDD579 pKa = 3.84LLQTEE584 pKa = 4.91DD585 pKa = 4.07SIKK588 pKa = 11.01GNTTEE593 pKa = 4.2VYY595 pKa = 9.97TGFIYY600 pKa = 10.85DD601 pKa = 3.74ADD603 pKa = 3.87GHH605 pKa = 6.51ISFYY609 pKa = 10.14EE610 pKa = 4.46HH611 pKa = 6.94IDD613 pKa = 3.45DD614 pKa = 5.24SVRR617 pKa = 11.84LYY619 pKa = 10.53IDD621 pKa = 3.54GVLVLSNDD629 pKa = 2.74SWEE632 pKa = 4.61ASSQTTDD639 pKa = 3.62LNLTPGTHH647 pKa = 6.53EE648 pKa = 3.7IEE650 pKa = 4.19LRR652 pKa = 11.84IGNADD657 pKa = 3.67GGSGAVDD664 pKa = 4.42GIGFGIDD671 pKa = 2.98VDD673 pKa = 3.83GGTNFVHH680 pKa = 7.4PSTLSEE686 pKa = 4.58SIFTSVGEE694 pKa = 4.19EE695 pKa = 3.89TGNPDD700 pKa = 4.41LEE702 pKa = 4.37QEE704 pKa = 3.85GDD706 pKa = 3.6IIVEE710 pKa = 4.05LEE712 pKa = 4.17SFVFTSTNGRR722 pKa = 11.84VGGDD726 pKa = 3.35SVEE729 pKa = 4.47GFSPTATGVNWVTNGDD745 pKa = 3.69YY746 pKa = 11.08GDD748 pKa = 3.86YY749 pKa = 9.75MVTFEE754 pKa = 5.2EE755 pKa = 4.89PGTYY759 pKa = 9.38SAYY762 pKa = 8.88ITISAANDD770 pKa = 3.45GSYY773 pKa = 10.64GARR776 pKa = 11.84VDD778 pKa = 3.47VDD780 pKa = 3.51GWPVAWGYY788 pKa = 11.05FGGTGSWDD796 pKa = 3.32VSSEE800 pKa = 3.57NLLYY804 pKa = 10.94GGTFVVEE811 pKa = 3.88QAGEE815 pKa = 3.97KK816 pKa = 9.34VVRR819 pKa = 11.84VEE821 pKa = 5.28AIGGSDD827 pKa = 3.53WQWSGDD833 pKa = 3.16RR834 pKa = 11.84VRR836 pKa = 11.84FTRR839 pKa = 11.84LGDD842 pKa = 3.31VTAIPSPIYY851 pKa = 10.84NPDD854 pKa = 3.21DD855 pKa = 3.96HH856 pKa = 7.09FVAEE860 pKa = 4.25IQGPQTDD867 pKa = 3.52VTYY870 pKa = 10.65LKK872 pKa = 10.73KK873 pKa = 10.31PVEE876 pKa = 3.9IPANKK881 pKa = 9.53KK882 pKa = 8.2VLKK885 pKa = 10.35SDD887 pKa = 2.47VWYY890 pKa = 8.63TYY892 pKa = 8.87PQNRR896 pKa = 11.84EE897 pKa = 3.72LEE899 pKa = 4.62GYY901 pKa = 10.75DD902 pKa = 3.25NFGATGAFWGHH913 pKa = 6.31PPEE916 pKa = 6.01HH917 pKa = 7.32DD918 pKa = 4.04FYY920 pKa = 11.46DD921 pKa = 3.9DD922 pKa = 3.6TVIMDD927 pKa = 4.26WAVDD931 pKa = 3.43AVYY934 pKa = 10.73AFQAEE939 pKa = 5.03GYY941 pKa = 8.67EE942 pKa = 4.05YY943 pKa = 9.77TARR946 pKa = 11.84GEE948 pKa = 4.11FDD950 pKa = 2.68WGYY953 pKa = 11.45GWFTEE958 pKa = 4.56YY959 pKa = 7.85TTNPQPHH966 pKa = 5.68YY967 pKa = 11.23VRR969 pKa = 11.84TLDD972 pKa = 3.67DD973 pKa = 3.98RR974 pKa = 11.84NVRR977 pKa = 11.84MTFMGYY983 pKa = 10.5LSHH986 pKa = 7.94DD987 pKa = 3.87GYY989 pKa = 11.54NNNWLSNHH997 pKa = 5.71SPAFVPFMKK1006 pKa = 10.46SQVDD1010 pKa = 3.78QILKK1014 pKa = 10.51ANPDD1018 pKa = 3.23KK1019 pKa = 11.52LMFDD1023 pKa = 3.57TQTNSTRR1030 pKa = 11.84STDD1033 pKa = 3.25MRR1035 pKa = 11.84DD1036 pKa = 3.24FGGDD1040 pKa = 3.54FSPYY1044 pKa = 9.96AMEE1047 pKa = 4.92NFRR1050 pKa = 11.84VWLSKK1055 pKa = 10.49KK1056 pKa = 10.39YY1057 pKa = 9.44STGEE1061 pKa = 3.73LAALGINDD1069 pKa = 4.36INSFDD1074 pKa = 3.68YY1075 pKa = 11.46GDD1077 pKa = 3.75FLRR1080 pKa = 11.84AQGVTHH1086 pKa = 6.5TSWSNAGDD1094 pKa = 3.76TLSGNIPLQEE1104 pKa = 4.06DD1105 pKa = 4.21YY1106 pKa = 10.91IYY1108 pKa = 10.85FNRR1111 pKa = 11.84DD1112 pKa = 1.96VWNQKK1117 pKa = 8.47FAEE1120 pKa = 4.14VLDD1123 pKa = 4.67YY1124 pKa = 10.77IRR1126 pKa = 11.84QQQPDD1131 pKa = 3.68IEE1133 pKa = 5.14IGASTHH1139 pKa = 5.75LFEE1142 pKa = 5.23SRR1144 pKa = 11.84GYY1146 pKa = 10.33VFNEE1150 pKa = 3.35NLTFLSGEE1158 pKa = 4.24LNLGARR1164 pKa = 11.84TTISEE1169 pKa = 4.63LPTNILVHH1177 pKa = 6.42LKK1179 pKa = 10.2GAQAVDD1185 pKa = 3.12KK1186 pKa = 9.61TLVYY1190 pKa = 10.12FPYY1193 pKa = 9.78PWEE1196 pKa = 4.08FDD1198 pKa = 3.43EE1199 pKa = 6.74LRR1201 pKa = 11.84LQDD1204 pKa = 3.35APRR1207 pKa = 11.84FGRR1210 pKa = 11.84GWVAQAYY1217 pKa = 9.84AYY1219 pKa = 10.24GGLFSIPANVWVGGEE1234 pKa = 3.93VWTWSPGADD1243 pKa = 3.33NYY1245 pKa = 11.19RR1246 pKa = 11.84DD1247 pKa = 2.94IYY1249 pKa = 11.15LFVRR1253 pKa = 11.84AQADD1257 pKa = 3.7LLDD1260 pKa = 5.57DD1261 pKa = 3.66YY1262 pKa = 11.46TSYY1265 pKa = 11.69SKK1267 pKa = 11.15VGLVHH1272 pKa = 7.3AMYY1275 pKa = 11.07SSMKK1279 pKa = 10.49AGFIDD1284 pKa = 4.46GGNQIQSSTKK1294 pKa = 10.39LLTEE1298 pKa = 4.09GNINFDD1304 pKa = 3.9LLVFGDD1310 pKa = 3.51EE1311 pKa = 4.94GYY1313 pKa = 9.94PVVPRR1318 pKa = 11.84PEE1320 pKa = 5.09DD1321 pKa = 3.49FDD1323 pKa = 4.58KK1324 pKa = 10.84FDD1326 pKa = 5.44HH1327 pKa = 6.72IFFDD1331 pKa = 4.82GDD1333 pKa = 3.54QQYY1336 pKa = 7.85LTAEE1340 pKa = 4.11QQALLDD1346 pKa = 3.79QQGDD1350 pKa = 3.81KK1351 pKa = 10.67VRR1353 pKa = 11.84HH1354 pKa = 5.17IGQRR1358 pKa = 11.84GTVSGIEE1365 pKa = 3.65ITVSINGTEE1374 pKa = 4.17SNEE1377 pKa = 4.15TVSAVSRR1384 pKa = 11.84IHH1386 pKa = 5.76EE1387 pKa = 4.15TDD1389 pKa = 2.75AAAPYY1394 pKa = 9.81VVHH1397 pKa = 6.67LVNRR1401 pKa = 11.84PFAGGVTPTLNNVEE1415 pKa = 4.2VAIPQSYY1422 pKa = 9.36FPEE1425 pKa = 4.52VVTGATLHH1433 pKa = 6.63LPDD1436 pKa = 4.49GTSTPLTLSTNADD1449 pKa = 3.39GDD1451 pKa = 4.0VVLPVNNLEE1460 pKa = 3.82VWGILEE1466 pKa = 4.3LAHH1469 pKa = 7.01
Molecular weight: 159.01 kDa
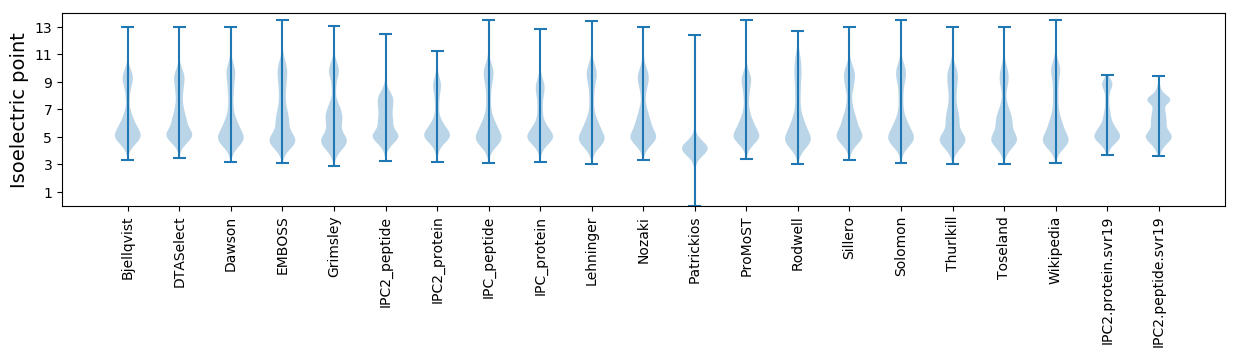
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0GDB0|A0A1I0GDB0_THASX Xaa-Pro dipeptidase OS=Thalassotalea agarivorans OX=349064 GN=pepQ PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
993003 |
27 |
3292 |
331.7 |
36.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.893 ± 0.044 | 0.925 ± 0.014 |
5.871 ± 0.042 | 6.102 ± 0.038 |
4.408 ± 0.03 | 6.679 ± 0.046 |
2.18 ± 0.024 | 6.527 ± 0.04 |
5.674 ± 0.041 | 9.815 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.438 ± 0.022 | 4.529 ± 0.031 |
3.798 ± 0.024 | 4.661 ± 0.041 |
3.992 ± 0.031 | 6.434 ± 0.034 |
5.482 ± 0.034 | 7.091 ± 0.04 |
1.217 ± 0.015 | 3.282 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
