
archaeon HR05
Taxonomy: cellular organisms; Archaea; unclassified Archaea
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
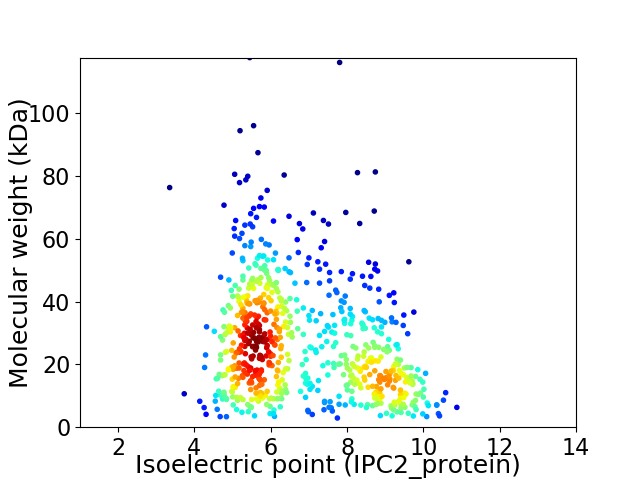
Virtual 2D-PAGE plot for 721 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5VBX8|A0A2H5VBX8_9ARCH Proteasome subunit beta OS=archaeon HR05 OX=2035441 GN=prcB_1 PE=3 SV=1
MM1 pKa = 7.85AGRR4 pKa = 11.84ASRR7 pKa = 11.84VRR9 pKa = 11.84GTTITVIATFLLLIIVLSTTITDD32 pKa = 3.62SLMTVYY38 pKa = 10.44SQEE41 pKa = 4.05QEE43 pKa = 3.68QTSAVEE49 pKa = 4.34CNGHH53 pKa = 6.5IIEE56 pKa = 5.84RR57 pKa = 11.84GDD59 pKa = 3.47FDD61 pKa = 4.28NDD63 pKa = 3.0EE64 pKa = 4.05VDD66 pKa = 3.43EE67 pKa = 4.6VKK69 pKa = 10.78VDD71 pKa = 3.37GTVYY75 pKa = 10.85EE76 pKa = 5.14DD77 pKa = 3.83GDD79 pKa = 5.09SIILDD84 pKa = 3.46GNTYY88 pKa = 5.77TTRR91 pKa = 11.84IVTVYY96 pKa = 10.44LFNGTNDD103 pKa = 2.94NDD105 pKa = 4.44FIVGRR110 pKa = 11.84NVSDD114 pKa = 4.23IIRR117 pKa = 11.84GKK119 pKa = 10.58EE120 pKa = 3.65GDD122 pKa = 3.75DD123 pKa = 4.04FIVGFGDD130 pKa = 3.38NDD132 pKa = 3.8SLYY135 pKa = 11.13GDD137 pKa = 4.57DD138 pKa = 6.25LFGEE142 pKa = 4.93DD143 pKa = 4.76GNDD146 pKa = 3.7TITGGNDD153 pKa = 3.11ILCGNAGYY161 pKa = 10.61DD162 pKa = 3.48HH163 pKa = 7.61LYY165 pKa = 10.98GDD167 pKa = 4.86MIGGGDD173 pKa = 3.94GNDD176 pKa = 4.03TISGGDD182 pKa = 3.67DD183 pKa = 2.84ILYY186 pKa = 10.64GGEE189 pKa = 4.53DD190 pKa = 3.57NDD192 pKa = 4.17EE193 pKa = 4.2LVGDD197 pKa = 5.1DD198 pKa = 4.26LSGWDD203 pKa = 3.97GDD205 pKa = 5.35DD206 pKa = 5.15NITGNNDD213 pKa = 2.91VLEE216 pKa = 4.94GGTGDD221 pKa = 3.32DD222 pKa = 3.31WLYY225 pKa = 11.76GDD227 pKa = 4.67MLHH230 pKa = 7.15GGVGSDD236 pKa = 4.09NITGGKK242 pKa = 7.28DD243 pKa = 3.01TLYY246 pKa = 10.93GGEE249 pKa = 4.24GNDD252 pKa = 3.57RR253 pKa = 11.84LFGDD257 pKa = 4.43SIGGGVGNNTITGGNDD273 pKa = 3.07TLDD276 pKa = 3.9GGDD279 pKa = 4.81GNDD282 pKa = 3.42EE283 pKa = 4.39LYY285 pKa = 11.14GDD287 pKa = 4.21GMEE290 pKa = 5.23GGDD293 pKa = 4.51GNDD296 pKa = 4.0TIKK299 pKa = 11.11GGADD303 pKa = 3.18TLDD306 pKa = 3.96GGDD309 pKa = 3.87GNDD312 pKa = 3.25WLIGDD317 pKa = 5.62RR318 pKa = 11.84IWGWDD323 pKa = 3.35GDD325 pKa = 4.16DD326 pKa = 4.5TISGGDD332 pKa = 3.33DD333 pKa = 3.15TLYY336 pKa = 11.22GGIGNDD342 pKa = 3.36HH343 pKa = 7.3LYY345 pKa = 11.32GDD347 pKa = 4.64MIGGEE352 pKa = 4.29DD353 pKa = 4.45GYY355 pKa = 10.34NTITGGKK362 pKa = 7.04DD363 pKa = 2.87TLYY366 pKa = 10.93GGEE369 pKa = 4.24GNDD372 pKa = 3.57RR373 pKa = 11.84LFGDD377 pKa = 4.43SIGGGVGNNTITGGNDD393 pKa = 3.07TLDD396 pKa = 3.9GGDD399 pKa = 4.81GNDD402 pKa = 3.42EE403 pKa = 4.39LYY405 pKa = 11.14GDD407 pKa = 4.21GMEE410 pKa = 5.23GGDD413 pKa = 4.51GNDD416 pKa = 4.0TIKK419 pKa = 11.11GGADD423 pKa = 3.18TLDD426 pKa = 3.96GGDD429 pKa = 3.87GNDD432 pKa = 3.25WLIGDD437 pKa = 5.62RR438 pKa = 11.84IWGWDD443 pKa = 3.35GDD445 pKa = 4.16DD446 pKa = 4.5TISGGDD452 pKa = 3.33DD453 pKa = 3.15TLYY456 pKa = 11.22GGIGNDD462 pKa = 3.36HH463 pKa = 7.3LYY465 pKa = 11.32GDD467 pKa = 4.64MIGGEE472 pKa = 4.29DD473 pKa = 4.45GYY475 pKa = 10.34NTITGGKK482 pKa = 7.04DD483 pKa = 2.87TLYY486 pKa = 10.93GGEE489 pKa = 4.24GNDD492 pKa = 3.57RR493 pKa = 11.84LFGDD497 pKa = 4.43SIGGGVGNNTITGGNDD513 pKa = 3.07TLDD516 pKa = 3.9GGDD519 pKa = 4.81GNDD522 pKa = 3.4EE523 pKa = 4.51LYY525 pKa = 11.25GDD527 pKa = 4.95GLFGWGGDD535 pKa = 3.38DD536 pKa = 3.25TAYY539 pKa = 10.78GGNDD543 pKa = 3.64TINAKK548 pKa = 10.67DD549 pKa = 3.57NADD552 pKa = 3.65NNDD555 pKa = 4.39NIDD558 pKa = 2.88GDD560 pKa = 4.48YY561 pKa = 10.86IVNAEE566 pKa = 4.32IATLDD571 pKa = 3.67NQDD574 pKa = 3.35ICASDD579 pKa = 4.18PDD581 pKa = 4.19PEE583 pKa = 4.66VNCEE587 pKa = 3.63YY588 pKa = 11.38DD589 pKa = 4.83DD590 pKa = 4.9IYY592 pKa = 11.61TLATLSTGTDD602 pKa = 3.5PVQLGEE608 pKa = 4.08PVQITFSSTVDD619 pKa = 3.37NPVKK623 pKa = 9.79ITSLTVTTPTDD634 pKa = 3.73SICTYY639 pKa = 10.12NSLPIIVPEE648 pKa = 4.35NGTFTATYY656 pKa = 8.44PADD659 pKa = 4.97FIGIDD664 pKa = 4.33CNTDD668 pKa = 2.83TAGTYY673 pKa = 7.39TVEE676 pKa = 5.16AITEE680 pKa = 4.12VGDD683 pKa = 4.6PITTNFTISFQVVPEE698 pKa = 5.0AIAGALGIVGTGFISMLLYY717 pKa = 10.58RR718 pKa = 11.84RR719 pKa = 11.84VSKK722 pKa = 11.07GKK724 pKa = 10.79GKK726 pKa = 10.34DD727 pKa = 3.08KK728 pKa = 11.38GKK730 pKa = 10.82DD731 pKa = 3.27NVASSS736 pKa = 3.46
MM1 pKa = 7.85AGRR4 pKa = 11.84ASRR7 pKa = 11.84VRR9 pKa = 11.84GTTITVIATFLLLIIVLSTTITDD32 pKa = 3.62SLMTVYY38 pKa = 10.44SQEE41 pKa = 4.05QEE43 pKa = 3.68QTSAVEE49 pKa = 4.34CNGHH53 pKa = 6.5IIEE56 pKa = 5.84RR57 pKa = 11.84GDD59 pKa = 3.47FDD61 pKa = 4.28NDD63 pKa = 3.0EE64 pKa = 4.05VDD66 pKa = 3.43EE67 pKa = 4.6VKK69 pKa = 10.78VDD71 pKa = 3.37GTVYY75 pKa = 10.85EE76 pKa = 5.14DD77 pKa = 3.83GDD79 pKa = 5.09SIILDD84 pKa = 3.46GNTYY88 pKa = 5.77TTRR91 pKa = 11.84IVTVYY96 pKa = 10.44LFNGTNDD103 pKa = 2.94NDD105 pKa = 4.44FIVGRR110 pKa = 11.84NVSDD114 pKa = 4.23IIRR117 pKa = 11.84GKK119 pKa = 10.58EE120 pKa = 3.65GDD122 pKa = 3.75DD123 pKa = 4.04FIVGFGDD130 pKa = 3.38NDD132 pKa = 3.8SLYY135 pKa = 11.13GDD137 pKa = 4.57DD138 pKa = 6.25LFGEE142 pKa = 4.93DD143 pKa = 4.76GNDD146 pKa = 3.7TITGGNDD153 pKa = 3.11ILCGNAGYY161 pKa = 10.61DD162 pKa = 3.48HH163 pKa = 7.61LYY165 pKa = 10.98GDD167 pKa = 4.86MIGGGDD173 pKa = 3.94GNDD176 pKa = 4.03TISGGDD182 pKa = 3.67DD183 pKa = 2.84ILYY186 pKa = 10.64GGEE189 pKa = 4.53DD190 pKa = 3.57NDD192 pKa = 4.17EE193 pKa = 4.2LVGDD197 pKa = 5.1DD198 pKa = 4.26LSGWDD203 pKa = 3.97GDD205 pKa = 5.35DD206 pKa = 5.15NITGNNDD213 pKa = 2.91VLEE216 pKa = 4.94GGTGDD221 pKa = 3.32DD222 pKa = 3.31WLYY225 pKa = 11.76GDD227 pKa = 4.67MLHH230 pKa = 7.15GGVGSDD236 pKa = 4.09NITGGKK242 pKa = 7.28DD243 pKa = 3.01TLYY246 pKa = 10.93GGEE249 pKa = 4.24GNDD252 pKa = 3.57RR253 pKa = 11.84LFGDD257 pKa = 4.43SIGGGVGNNTITGGNDD273 pKa = 3.07TLDD276 pKa = 3.9GGDD279 pKa = 4.81GNDD282 pKa = 3.42EE283 pKa = 4.39LYY285 pKa = 11.14GDD287 pKa = 4.21GMEE290 pKa = 5.23GGDD293 pKa = 4.51GNDD296 pKa = 4.0TIKK299 pKa = 11.11GGADD303 pKa = 3.18TLDD306 pKa = 3.96GGDD309 pKa = 3.87GNDD312 pKa = 3.25WLIGDD317 pKa = 5.62RR318 pKa = 11.84IWGWDD323 pKa = 3.35GDD325 pKa = 4.16DD326 pKa = 4.5TISGGDD332 pKa = 3.33DD333 pKa = 3.15TLYY336 pKa = 11.22GGIGNDD342 pKa = 3.36HH343 pKa = 7.3LYY345 pKa = 11.32GDD347 pKa = 4.64MIGGEE352 pKa = 4.29DD353 pKa = 4.45GYY355 pKa = 10.34NTITGGKK362 pKa = 7.04DD363 pKa = 2.87TLYY366 pKa = 10.93GGEE369 pKa = 4.24GNDD372 pKa = 3.57RR373 pKa = 11.84LFGDD377 pKa = 4.43SIGGGVGNNTITGGNDD393 pKa = 3.07TLDD396 pKa = 3.9GGDD399 pKa = 4.81GNDD402 pKa = 3.42EE403 pKa = 4.39LYY405 pKa = 11.14GDD407 pKa = 4.21GMEE410 pKa = 5.23GGDD413 pKa = 4.51GNDD416 pKa = 4.0TIKK419 pKa = 11.11GGADD423 pKa = 3.18TLDD426 pKa = 3.96GGDD429 pKa = 3.87GNDD432 pKa = 3.25WLIGDD437 pKa = 5.62RR438 pKa = 11.84IWGWDD443 pKa = 3.35GDD445 pKa = 4.16DD446 pKa = 4.5TISGGDD452 pKa = 3.33DD453 pKa = 3.15TLYY456 pKa = 11.22GGIGNDD462 pKa = 3.36HH463 pKa = 7.3LYY465 pKa = 11.32GDD467 pKa = 4.64MIGGEE472 pKa = 4.29DD473 pKa = 4.45GYY475 pKa = 10.34NTITGGKK482 pKa = 7.04DD483 pKa = 2.87TLYY486 pKa = 10.93GGEE489 pKa = 4.24GNDD492 pKa = 3.57RR493 pKa = 11.84LFGDD497 pKa = 4.43SIGGGVGNNTITGGNDD513 pKa = 3.07TLDD516 pKa = 3.9GGDD519 pKa = 4.81GNDD522 pKa = 3.4EE523 pKa = 4.51LYY525 pKa = 11.25GDD527 pKa = 4.95GLFGWGGDD535 pKa = 3.38DD536 pKa = 3.25TAYY539 pKa = 10.78GGNDD543 pKa = 3.64TINAKK548 pKa = 10.67DD549 pKa = 3.57NADD552 pKa = 3.65NNDD555 pKa = 4.39NIDD558 pKa = 2.88GDD560 pKa = 4.48YY561 pKa = 10.86IVNAEE566 pKa = 4.32IATLDD571 pKa = 3.67NQDD574 pKa = 3.35ICASDD579 pKa = 4.18PDD581 pKa = 4.19PEE583 pKa = 4.66VNCEE587 pKa = 3.63YY588 pKa = 11.38DD589 pKa = 4.83DD590 pKa = 4.9IYY592 pKa = 11.61TLATLSTGTDD602 pKa = 3.5PVQLGEE608 pKa = 4.08PVQITFSSTVDD619 pKa = 3.37NPVKK623 pKa = 9.79ITSLTVTTPTDD634 pKa = 3.73SICTYY639 pKa = 10.12NSLPIIVPEE648 pKa = 4.35NGTFTATYY656 pKa = 8.44PADD659 pKa = 4.97FIGIDD664 pKa = 4.33CNTDD668 pKa = 2.83TAGTYY673 pKa = 7.39TVEE676 pKa = 5.16AITEE680 pKa = 4.12VGDD683 pKa = 4.6PITTNFTISFQVVPEE698 pKa = 5.0AIAGALGIVGTGFISMLLYY717 pKa = 10.58RR718 pKa = 11.84RR719 pKa = 11.84VSKK722 pKa = 11.07GKK724 pKa = 10.79GKK726 pKa = 10.34DD727 pKa = 3.08KK728 pKa = 11.38GKK730 pKa = 10.82DD731 pKa = 3.27NVASSS736 pKa = 3.46
Molecular weight: 76.28 kDa
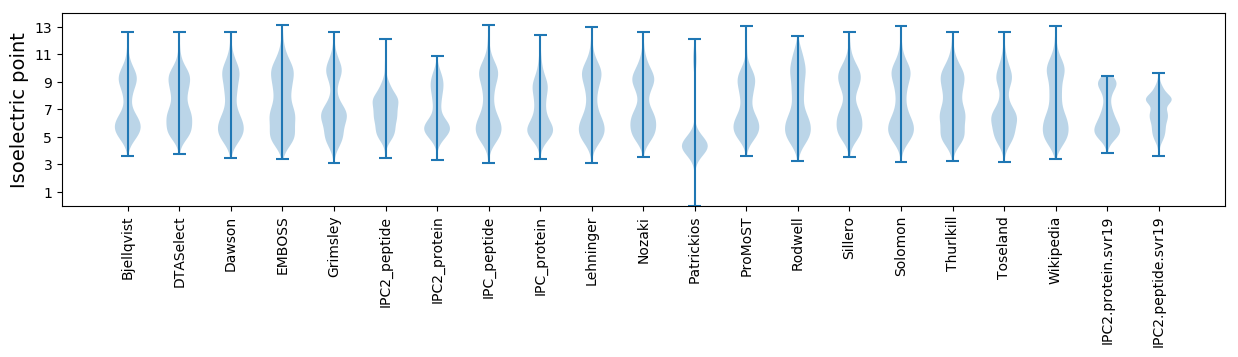
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5VCK6|A0A2H5VCK6_9ARCH Hypoxic response protein 1 OS=archaeon HR05 OX=2035441 GN=hrp1 PE=4 SV=1
MM1 pKa = 7.81GSRR4 pKa = 11.84KK5 pKa = 7.97TKK7 pKa = 8.99PRR9 pKa = 11.84KK10 pKa = 8.81NRR12 pKa = 11.84LMKK15 pKa = 9.39KK16 pKa = 7.91TKK18 pKa = 10.09QNSPVPLWVILKK30 pKa = 7.0TNRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84NPQQRR42 pKa = 11.84HH43 pKa = 3.83WRR45 pKa = 11.84RR46 pKa = 11.84SSVDD50 pKa = 3.09VGG52 pKa = 3.49
MM1 pKa = 7.81GSRR4 pKa = 11.84KK5 pKa = 7.97TKK7 pKa = 8.99PRR9 pKa = 11.84KK10 pKa = 8.81NRR12 pKa = 11.84LMKK15 pKa = 9.39KK16 pKa = 7.91TKK18 pKa = 10.09QNSPVPLWVILKK30 pKa = 7.0TNRR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84RR37 pKa = 11.84NPQQRR42 pKa = 11.84HH43 pKa = 3.83WRR45 pKa = 11.84RR46 pKa = 11.84SSVDD50 pKa = 3.09VGG52 pKa = 3.49
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
177987 |
29 |
1023 |
246.9 |
27.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.739 ± 0.097 | 1.087 ± 0.039 |
6.01 ± 0.089 | 6.504 ± 0.092 |
3.1 ± 0.065 | 7.024 ± 0.1 |
1.965 ± 0.042 | 8.352 ± 0.088 |
5.748 ± 0.082 | 9.257 ± 0.095 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.488 ± 0.049 | 3.999 ± 0.068 |
3.381 ± 0.057 | 1.868 ± 0.042 |
6.553 ± 0.084 | 7.09 ± 0.091 |
4.21 ± 0.062 | 7.907 ± 0.078 |
0.827 ± 0.032 | 3.89 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
