
Beihai sobemo-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
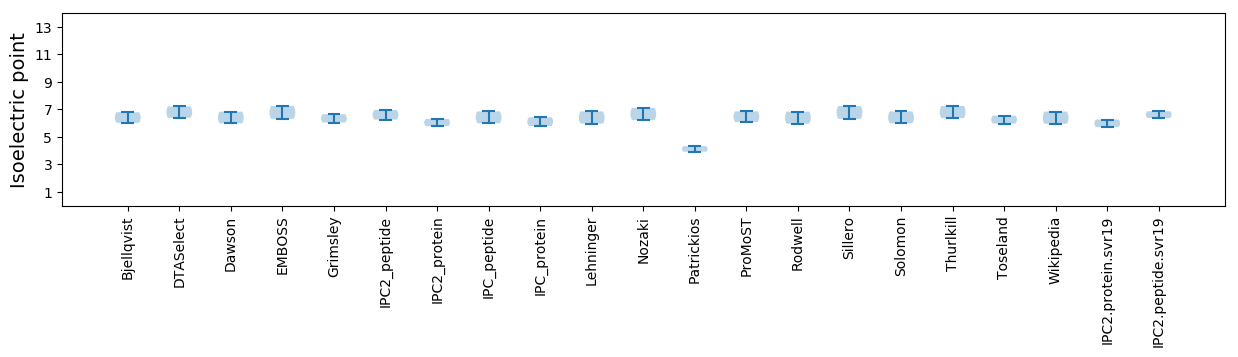
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
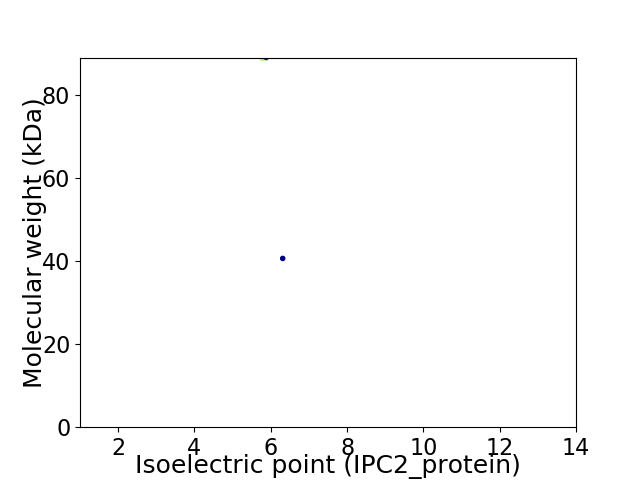
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE56|A0A1L3KE56_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 15 OX=1922686 PE=4 SV=1
MM1 pKa = 7.48SFNGVLPNQFHH12 pKa = 6.8LEE14 pKa = 4.09EE15 pKa = 4.81HH16 pKa = 6.73LSPYY20 pKa = 10.29TNYY23 pKa = 10.11QGSAPPMDD31 pKa = 4.22FHH33 pKa = 7.08PLGIKK38 pKa = 9.44TIEE41 pKa = 4.08LVHH44 pKa = 7.08ILLASLILLTMGLMCALLRR63 pKa = 11.84DD64 pKa = 4.41WFATTPLRR72 pKa = 11.84DD73 pKa = 3.24LLSRR77 pKa = 11.84KK78 pKa = 9.17RR79 pKa = 11.84RR80 pKa = 11.84PKK82 pKa = 10.72HH83 pKa = 5.86NIPDD87 pKa = 3.5ATEE90 pKa = 4.37DD91 pKa = 3.76EE92 pKa = 4.94EE93 pKa = 5.01EE94 pKa = 4.11EE95 pKa = 4.16QPLVVDD101 pKa = 4.44EE102 pKa = 4.89CPKK105 pKa = 10.42CRR107 pKa = 11.84PQQSPQPEE115 pKa = 4.4SNDD118 pKa = 3.21YY119 pKa = 8.2FTKK122 pKa = 10.67CKK124 pKa = 10.28LPKK127 pKa = 9.75EE128 pKa = 4.05QVILWRR134 pKa = 11.84TCCKK138 pKa = 10.37SDD140 pKa = 5.39DD141 pKa = 4.01PAKK144 pKa = 10.54PPLTTHH150 pKa = 6.3VGQAFFIAKK159 pKa = 7.85DD160 pKa = 3.62TLLTTAHH167 pKa = 6.57SIKK170 pKa = 10.44PGEE173 pKa = 4.35KK174 pKa = 9.45YY175 pKa = 10.78VISKK179 pKa = 9.45SQIGRR184 pKa = 11.84AFPVLSVQYY193 pKa = 7.98PTGPAKK199 pKa = 10.46DD200 pKa = 3.67VIYY203 pKa = 10.74LKK205 pKa = 10.73VEE207 pKa = 3.75NRR209 pKa = 11.84AFTQLEE215 pKa = 3.89ASVAKK220 pKa = 10.13IASEE224 pKa = 4.07YY225 pKa = 11.0LPGDD229 pKa = 3.75TVSVCGGGLEE239 pKa = 4.18PQEE242 pKa = 4.46SSDD245 pKa = 4.78RR246 pKa = 11.84ALQSIGNLEE255 pKa = 4.11PWEE258 pKa = 4.37GAAGILISAHH268 pKa = 5.74NCNTLKK274 pKa = 10.66GFSGAPIFCGNKK286 pKa = 8.26VVAMHH291 pKa = 6.32NQTQTDD297 pKa = 3.78MFNIATNLWPVLGTKK312 pKa = 10.05CQISPQFEE320 pKa = 4.38SSEE323 pKa = 3.66MDD325 pKa = 3.25ANFFRR330 pKa = 11.84HH331 pKa = 6.92DD332 pKa = 3.47DD333 pKa = 3.77TRR335 pKa = 11.84ILEE338 pKa = 4.42YY339 pKa = 11.03DD340 pKa = 3.44DD341 pKa = 3.58MVYY344 pKa = 10.71VSKK347 pKa = 11.02AGRR350 pKa = 11.84FYY352 pKa = 11.1NIDD355 pKa = 3.26RR356 pKa = 11.84QVYY359 pKa = 9.14QKK361 pKa = 10.93LKK363 pKa = 10.48QKK365 pKa = 9.78MPNTTDD371 pKa = 3.03SLYY374 pKa = 11.02VPPKK378 pKa = 8.88EE379 pKa = 4.28NAWADD384 pKa = 3.34GGEE387 pKa = 4.22EE388 pKa = 3.94IDD390 pKa = 4.47FNSPLVFEE398 pKa = 5.2SSNALPTPLTPLNVDD413 pKa = 3.76AVRR416 pKa = 11.84QANVAITQVNNAYY429 pKa = 9.88FALRR433 pKa = 11.84TMAMTTSRR441 pKa = 11.84LLNLALSQKK450 pKa = 10.54DD451 pKa = 3.53EE452 pKa = 4.44EE453 pKa = 4.83GAKK456 pKa = 10.36SLANNFTYY464 pKa = 10.53LSKK467 pKa = 10.86QLAPEE472 pKa = 4.25SFKK475 pKa = 10.26QTIRR479 pKa = 11.84DD480 pKa = 3.95AIDD483 pKa = 3.29NFSYY487 pKa = 9.9ATMLSEE493 pKa = 5.02QDD495 pKa = 3.02EE496 pKa = 4.47KK497 pKa = 11.35LAKK500 pKa = 9.36KK501 pKa = 8.48WKK503 pKa = 8.78KK504 pKa = 10.22RR505 pKa = 11.84RR506 pKa = 11.84DD507 pKa = 3.54AGDD510 pKa = 3.71PDD512 pKa = 5.23AEE514 pKa = 4.52FKK516 pKa = 11.01QITDD520 pKa = 3.9GYY522 pKa = 8.99IAEE525 pKa = 4.45IDD527 pKa = 3.89TYY529 pKa = 11.34SKK531 pKa = 11.31ALAGIPKK538 pKa = 10.06RR539 pKa = 11.84LNKK542 pKa = 9.91NDD544 pKa = 4.57LKK546 pKa = 10.83IQVPPEE552 pKa = 3.65PEE554 pKa = 3.76FNDD557 pKa = 3.55RR558 pKa = 11.84EE559 pKa = 4.01NAFFLKK565 pKa = 10.73YY566 pKa = 10.41GINFANPEE574 pKa = 4.42EE575 pKa = 4.34ILKK578 pKa = 10.8APAIEE583 pKa = 4.3LPKK586 pKa = 10.43KK587 pKa = 10.18PEE589 pKa = 3.67EE590 pKa = 4.26SEE592 pKa = 4.18PKK594 pKa = 10.58VILAPQRR601 pKa = 11.84PTQEE605 pKa = 3.84IRR607 pKa = 11.84NKK609 pKa = 5.46TTKK612 pKa = 10.48GKK614 pKa = 10.36LSKK617 pKa = 9.67TPNTSSVQVLNTPSAPSKK635 pKa = 10.59EE636 pKa = 4.19MEE638 pKa = 4.62AKK640 pKa = 10.49LVDD643 pKa = 3.77HH644 pKa = 6.3EE645 pKa = 5.0EE646 pKa = 4.13KK647 pKa = 10.86LKK649 pKa = 10.81SAKK652 pKa = 9.15QSKK655 pKa = 9.38LQEE658 pKa = 4.31EE659 pKa = 4.89CSGNCLTAQTGLEE672 pKa = 4.02SSIAQLTSLIQLLTTTIAASQLTSPPQTPSSKK704 pKa = 10.49LSPTTQKK711 pKa = 10.6KK712 pKa = 7.23PQGMSSLQAQPTQKK726 pKa = 10.11QSQLVSEE733 pKa = 4.86CTLSTIDD740 pKa = 4.98EE741 pKa = 4.21PRR743 pKa = 11.84KK744 pKa = 9.78DD745 pKa = 4.23LKK747 pKa = 11.26SLMQLQSEE755 pKa = 5.08GCRR758 pKa = 11.84RR759 pKa = 11.84DD760 pKa = 3.32MLSNLLRR767 pKa = 11.84PGVQCIKK774 pKa = 9.81ITGEE778 pKa = 4.18TIEE781 pKa = 4.73PCQDD785 pKa = 3.2SKK787 pKa = 11.71GSEE790 pKa = 3.95PSSLKK795 pKa = 8.21TTTDD799 pKa = 2.98AMCC802 pKa = 4.7
MM1 pKa = 7.48SFNGVLPNQFHH12 pKa = 6.8LEE14 pKa = 4.09EE15 pKa = 4.81HH16 pKa = 6.73LSPYY20 pKa = 10.29TNYY23 pKa = 10.11QGSAPPMDD31 pKa = 4.22FHH33 pKa = 7.08PLGIKK38 pKa = 9.44TIEE41 pKa = 4.08LVHH44 pKa = 7.08ILLASLILLTMGLMCALLRR63 pKa = 11.84DD64 pKa = 4.41WFATTPLRR72 pKa = 11.84DD73 pKa = 3.24LLSRR77 pKa = 11.84KK78 pKa = 9.17RR79 pKa = 11.84RR80 pKa = 11.84PKK82 pKa = 10.72HH83 pKa = 5.86NIPDD87 pKa = 3.5ATEE90 pKa = 4.37DD91 pKa = 3.76EE92 pKa = 4.94EE93 pKa = 5.01EE94 pKa = 4.11EE95 pKa = 4.16QPLVVDD101 pKa = 4.44EE102 pKa = 4.89CPKK105 pKa = 10.42CRR107 pKa = 11.84PQQSPQPEE115 pKa = 4.4SNDD118 pKa = 3.21YY119 pKa = 8.2FTKK122 pKa = 10.67CKK124 pKa = 10.28LPKK127 pKa = 9.75EE128 pKa = 4.05QVILWRR134 pKa = 11.84TCCKK138 pKa = 10.37SDD140 pKa = 5.39DD141 pKa = 4.01PAKK144 pKa = 10.54PPLTTHH150 pKa = 6.3VGQAFFIAKK159 pKa = 7.85DD160 pKa = 3.62TLLTTAHH167 pKa = 6.57SIKK170 pKa = 10.44PGEE173 pKa = 4.35KK174 pKa = 9.45YY175 pKa = 10.78VISKK179 pKa = 9.45SQIGRR184 pKa = 11.84AFPVLSVQYY193 pKa = 7.98PTGPAKK199 pKa = 10.46DD200 pKa = 3.67VIYY203 pKa = 10.74LKK205 pKa = 10.73VEE207 pKa = 3.75NRR209 pKa = 11.84AFTQLEE215 pKa = 3.89ASVAKK220 pKa = 10.13IASEE224 pKa = 4.07YY225 pKa = 11.0LPGDD229 pKa = 3.75TVSVCGGGLEE239 pKa = 4.18PQEE242 pKa = 4.46SSDD245 pKa = 4.78RR246 pKa = 11.84ALQSIGNLEE255 pKa = 4.11PWEE258 pKa = 4.37GAAGILISAHH268 pKa = 5.74NCNTLKK274 pKa = 10.66GFSGAPIFCGNKK286 pKa = 8.26VVAMHH291 pKa = 6.32NQTQTDD297 pKa = 3.78MFNIATNLWPVLGTKK312 pKa = 10.05CQISPQFEE320 pKa = 4.38SSEE323 pKa = 3.66MDD325 pKa = 3.25ANFFRR330 pKa = 11.84HH331 pKa = 6.92DD332 pKa = 3.47DD333 pKa = 3.77TRR335 pKa = 11.84ILEE338 pKa = 4.42YY339 pKa = 11.03DD340 pKa = 3.44DD341 pKa = 3.58MVYY344 pKa = 10.71VSKK347 pKa = 11.02AGRR350 pKa = 11.84FYY352 pKa = 11.1NIDD355 pKa = 3.26RR356 pKa = 11.84QVYY359 pKa = 9.14QKK361 pKa = 10.93LKK363 pKa = 10.48QKK365 pKa = 9.78MPNTTDD371 pKa = 3.03SLYY374 pKa = 11.02VPPKK378 pKa = 8.88EE379 pKa = 4.28NAWADD384 pKa = 3.34GGEE387 pKa = 4.22EE388 pKa = 3.94IDD390 pKa = 4.47FNSPLVFEE398 pKa = 5.2SSNALPTPLTPLNVDD413 pKa = 3.76AVRR416 pKa = 11.84QANVAITQVNNAYY429 pKa = 9.88FALRR433 pKa = 11.84TMAMTTSRR441 pKa = 11.84LLNLALSQKK450 pKa = 10.54DD451 pKa = 3.53EE452 pKa = 4.44EE453 pKa = 4.83GAKK456 pKa = 10.36SLANNFTYY464 pKa = 10.53LSKK467 pKa = 10.86QLAPEE472 pKa = 4.25SFKK475 pKa = 10.26QTIRR479 pKa = 11.84DD480 pKa = 3.95AIDD483 pKa = 3.29NFSYY487 pKa = 9.9ATMLSEE493 pKa = 5.02QDD495 pKa = 3.02EE496 pKa = 4.47KK497 pKa = 11.35LAKK500 pKa = 9.36KK501 pKa = 8.48WKK503 pKa = 8.78KK504 pKa = 10.22RR505 pKa = 11.84RR506 pKa = 11.84DD507 pKa = 3.54AGDD510 pKa = 3.71PDD512 pKa = 5.23AEE514 pKa = 4.52FKK516 pKa = 11.01QITDD520 pKa = 3.9GYY522 pKa = 8.99IAEE525 pKa = 4.45IDD527 pKa = 3.89TYY529 pKa = 11.34SKK531 pKa = 11.31ALAGIPKK538 pKa = 10.06RR539 pKa = 11.84LNKK542 pKa = 9.91NDD544 pKa = 4.57LKK546 pKa = 10.83IQVPPEE552 pKa = 3.65PEE554 pKa = 3.76FNDD557 pKa = 3.55RR558 pKa = 11.84EE559 pKa = 4.01NAFFLKK565 pKa = 10.73YY566 pKa = 10.41GINFANPEE574 pKa = 4.42EE575 pKa = 4.34ILKK578 pKa = 10.8APAIEE583 pKa = 4.3LPKK586 pKa = 10.43KK587 pKa = 10.18PEE589 pKa = 3.67EE590 pKa = 4.26SEE592 pKa = 4.18PKK594 pKa = 10.58VILAPQRR601 pKa = 11.84PTQEE605 pKa = 3.84IRR607 pKa = 11.84NKK609 pKa = 5.46TTKK612 pKa = 10.48GKK614 pKa = 10.36LSKK617 pKa = 9.67TPNTSSVQVLNTPSAPSKK635 pKa = 10.59EE636 pKa = 4.19MEE638 pKa = 4.62AKK640 pKa = 10.49LVDD643 pKa = 3.77HH644 pKa = 6.3EE645 pKa = 5.0EE646 pKa = 4.13KK647 pKa = 10.86LKK649 pKa = 10.81SAKK652 pKa = 9.15QSKK655 pKa = 9.38LQEE658 pKa = 4.31EE659 pKa = 4.89CSGNCLTAQTGLEE672 pKa = 4.02SSIAQLTSLIQLLTTTIAASQLTSPPQTPSSKK704 pKa = 10.49LSPTTQKK711 pKa = 10.6KK712 pKa = 7.23PQGMSSLQAQPTQKK726 pKa = 10.11QSQLVSEE733 pKa = 4.86CTLSTIDD740 pKa = 4.98EE741 pKa = 4.21PRR743 pKa = 11.84KK744 pKa = 9.78DD745 pKa = 4.23LKK747 pKa = 11.26SLMQLQSEE755 pKa = 5.08GCRR758 pKa = 11.84RR759 pKa = 11.84DD760 pKa = 3.32MLSNLLRR767 pKa = 11.84PGVQCIKK774 pKa = 9.81ITGEE778 pKa = 4.18TIEE781 pKa = 4.73PCQDD785 pKa = 3.2SKK787 pKa = 11.71GSEE790 pKa = 3.95PSSLKK795 pKa = 8.21TTTDD799 pKa = 2.98AMCC802 pKa = 4.7
Molecular weight: 89.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE56|A0A1L3KE56_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 15 OX=1922686 PE=4 SV=1
MM1 pKa = 7.49SGLEE5 pKa = 3.93GFRR8 pKa = 11.84AFISEE13 pKa = 4.12NHH15 pKa = 7.0DD16 pKa = 3.09GRR18 pKa = 11.84HH19 pKa = 4.45VLEE22 pKa = 4.45PTSTPGYY29 pKa = 9.52PWVQLGNSNAEE40 pKa = 4.14CFGYY44 pKa = 10.75VPGLGYY50 pKa = 11.0VGDD53 pKa = 4.28KK54 pKa = 11.01VADD57 pKa = 4.51LYY59 pKa = 11.04QAVRR63 pKa = 11.84QRR65 pKa = 11.84LYY67 pKa = 10.62TLEE70 pKa = 4.18TTPKK74 pKa = 9.17IDD76 pKa = 4.95NIFLFIKK83 pKa = 10.29QEE85 pKa = 3.75PHH87 pKa = 6.53KK88 pKa = 9.82PAKK91 pKa = 9.42AQQRR95 pKa = 11.84RR96 pKa = 11.84WRR98 pKa = 11.84LISGIGATDD107 pKa = 3.36QLIAEE112 pKa = 4.29MLFRR116 pKa = 11.84PFLDD120 pKa = 2.92EE121 pKa = 4.0MTNNPCRR128 pKa = 11.84YY129 pKa = 9.46GMAVGWAPVLEE140 pKa = 4.12NAIPFIMSHH149 pKa = 6.47LSGLTVCADD158 pKa = 3.27KK159 pKa = 11.25SAWDD163 pKa = 3.46WTVQPWLCDD172 pKa = 3.41YY173 pKa = 8.59MKK175 pKa = 11.0SFLSKK180 pKa = 10.43LCPGVSTVVQNHH192 pKa = 5.03VEE194 pKa = 3.72AVMGKK199 pKa = 8.06KK200 pKa = 8.08TFQWPGGKK208 pKa = 8.45YY209 pKa = 8.81HH210 pKa = 6.47WDD212 pKa = 3.14KK213 pKa = 11.46RR214 pKa = 11.84GVFCSGWKK222 pKa = 8.38LTITGNSIMQHH233 pKa = 5.38ACHH236 pKa = 6.52RR237 pKa = 11.84MAGGTGNMQANGDD250 pKa = 3.97DD251 pKa = 3.71TCQAEE256 pKa = 4.57EE257 pKa = 4.18PPEE260 pKa = 3.77YY261 pKa = 10.05WEE263 pKa = 4.28NLRR266 pKa = 11.84KK267 pKa = 9.95LGPIIKK273 pKa = 9.74EE274 pKa = 3.54IRR276 pKa = 11.84RR277 pKa = 11.84DD278 pKa = 3.58GTIEE282 pKa = 3.94FCGLEE287 pKa = 3.95ITPDD291 pKa = 3.46GYY293 pKa = 11.69YY294 pKa = 10.29PMYY297 pKa = 9.82GAKK300 pKa = 8.37HH301 pKa = 5.23TFVLTHH307 pKa = 6.91LKK309 pKa = 10.24PKK311 pKa = 10.04IARR314 pKa = 11.84EE315 pKa = 3.97VLLSYY320 pKa = 10.61QYY322 pKa = 11.12LYY324 pKa = 10.42PFEE327 pKa = 4.54PEE329 pKa = 3.62KK330 pKa = 10.87LAAIQAWIASLGFEE344 pKa = 4.88KK345 pKa = 10.77DD346 pKa = 3.28LTPPEE351 pKa = 4.26VLRR354 pKa = 11.84GVPLGVII361 pKa = 4.07
MM1 pKa = 7.49SGLEE5 pKa = 3.93GFRR8 pKa = 11.84AFISEE13 pKa = 4.12NHH15 pKa = 7.0DD16 pKa = 3.09GRR18 pKa = 11.84HH19 pKa = 4.45VLEE22 pKa = 4.45PTSTPGYY29 pKa = 9.52PWVQLGNSNAEE40 pKa = 4.14CFGYY44 pKa = 10.75VPGLGYY50 pKa = 11.0VGDD53 pKa = 4.28KK54 pKa = 11.01VADD57 pKa = 4.51LYY59 pKa = 11.04QAVRR63 pKa = 11.84QRR65 pKa = 11.84LYY67 pKa = 10.62TLEE70 pKa = 4.18TTPKK74 pKa = 9.17IDD76 pKa = 4.95NIFLFIKK83 pKa = 10.29QEE85 pKa = 3.75PHH87 pKa = 6.53KK88 pKa = 9.82PAKK91 pKa = 9.42AQQRR95 pKa = 11.84RR96 pKa = 11.84WRR98 pKa = 11.84LISGIGATDD107 pKa = 3.36QLIAEE112 pKa = 4.29MLFRR116 pKa = 11.84PFLDD120 pKa = 2.92EE121 pKa = 4.0MTNNPCRR128 pKa = 11.84YY129 pKa = 9.46GMAVGWAPVLEE140 pKa = 4.12NAIPFIMSHH149 pKa = 6.47LSGLTVCADD158 pKa = 3.27KK159 pKa = 11.25SAWDD163 pKa = 3.46WTVQPWLCDD172 pKa = 3.41YY173 pKa = 8.59MKK175 pKa = 11.0SFLSKK180 pKa = 10.43LCPGVSTVVQNHH192 pKa = 5.03VEE194 pKa = 3.72AVMGKK199 pKa = 8.06KK200 pKa = 8.08TFQWPGGKK208 pKa = 8.45YY209 pKa = 8.81HH210 pKa = 6.47WDD212 pKa = 3.14KK213 pKa = 11.46RR214 pKa = 11.84GVFCSGWKK222 pKa = 8.38LTITGNSIMQHH233 pKa = 5.38ACHH236 pKa = 6.52RR237 pKa = 11.84MAGGTGNMQANGDD250 pKa = 3.97DD251 pKa = 3.71TCQAEE256 pKa = 4.57EE257 pKa = 4.18PPEE260 pKa = 3.77YY261 pKa = 10.05WEE263 pKa = 4.28NLRR266 pKa = 11.84KK267 pKa = 9.95LGPIIKK273 pKa = 9.74EE274 pKa = 3.54IRR276 pKa = 11.84RR277 pKa = 11.84DD278 pKa = 3.58GTIEE282 pKa = 3.94FCGLEE287 pKa = 3.95ITPDD291 pKa = 3.46GYY293 pKa = 11.69YY294 pKa = 10.29PMYY297 pKa = 9.82GAKK300 pKa = 8.37HH301 pKa = 5.23TFVLTHH307 pKa = 6.91LKK309 pKa = 10.24PKK311 pKa = 10.04IARR314 pKa = 11.84EE315 pKa = 3.97VLLSYY320 pKa = 10.61QYY322 pKa = 11.12LYY324 pKa = 10.42PFEE327 pKa = 4.54PEE329 pKa = 3.62KK330 pKa = 10.87LAAIQAWIASLGFEE344 pKa = 4.88KK345 pKa = 10.77DD346 pKa = 3.28LTPPEE351 pKa = 4.26VLRR354 pKa = 11.84GVPLGVII361 pKa = 4.07
Molecular weight: 40.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1163 |
361 |
802 |
581.5 |
64.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.137 ± 0.265 | 2.236 ± 0.14 |
4.815 ± 0.358 | 6.707 ± 0.332 |
3.611 ± 0.295 | 5.847 ± 1.788 |
1.806 ± 0.523 | 5.245 ± 0.16 |
7.051 ± 0.82 | 9.63 ± 0.416 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.408 ± 0.347 | 4.299 ± 0.529 |
7.309 ± 0.208 | 5.503 ± 0.731 |
3.783 ± 0.352 | 6.879 ± 1.478 |
6.793 ± 0.68 | 4.643 ± 0.637 |
1.462 ± 0.86 | 2.837 ± 0.715 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
