
Euzebyella marina
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Euzebyella
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
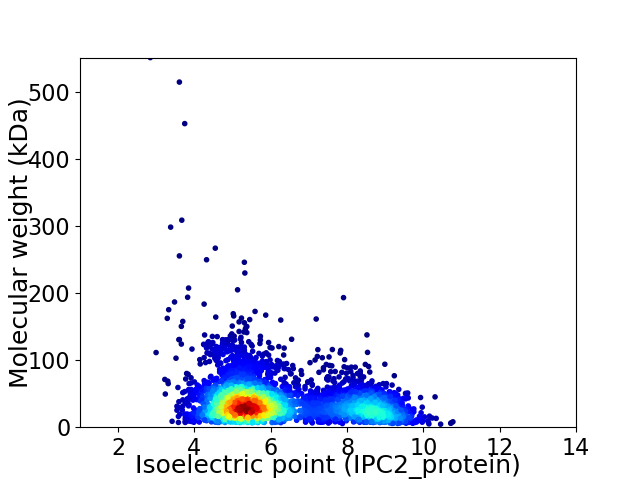
Virtual 2D-PAGE plot for 3952 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G2L9B8|A0A3G2L9B8_9FLAO Putative fluoride ion transporter CrcB OS=Euzebyella marina OX=1761453 GN=crcB PE=3 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84FLLALSLVLLFGLQSQAQNSPDD24 pKa = 3.7CRR26 pKa = 11.84SAIPVCADD34 pKa = 3.11APIMGRR40 pKa = 11.84ADD42 pKa = 3.34GGGDD46 pKa = 3.23IDD48 pKa = 5.85DD49 pKa = 5.34FDD51 pKa = 5.93PEE53 pKa = 4.66VITEE57 pKa = 4.4TGCLEE62 pKa = 4.24KK63 pKa = 11.09GSVSNANIEE72 pKa = 4.6NNTSWYY78 pKa = 9.78VFRR81 pKa = 11.84AGTGGQVGFDD91 pKa = 3.54IEE93 pKa = 4.71ALPLPGEE100 pKa = 4.31TTITAEE106 pKa = 3.5WDD108 pKa = 3.61FAVYY112 pKa = 10.33GPDD115 pKa = 3.48VACGEE120 pKa = 4.24ISSGQAQPIRR130 pKa = 11.84CNYY133 pKa = 7.81EE134 pKa = 3.97VNGTRR139 pKa = 11.84YY140 pKa = 8.26TGIGVNPEE148 pKa = 4.11DD149 pKa = 4.1GQAGVPSTTQNLNTYY164 pKa = 9.36DD165 pKa = 3.25EE166 pKa = 4.57WLDD169 pKa = 3.61VLPGEE174 pKa = 4.81VYY176 pKa = 10.98YY177 pKa = 10.27ILINNFSNNTNNDD190 pKa = 3.41PEE192 pKa = 4.34EE193 pKa = 4.78FILTFTGNSVEE204 pKa = 4.38ADD206 pKa = 3.15QNTALDD212 pKa = 3.73CTLRR216 pKa = 11.84DD217 pKa = 3.55EE218 pKa = 5.64FLGLDD223 pKa = 3.36INACEE228 pKa = 4.27GDD230 pKa = 3.67PDD232 pKa = 3.97IEE234 pKa = 4.12LVALNSPAGPDD245 pKa = 3.12IANVAWSVDD254 pKa = 3.85FDD256 pKa = 5.18DD257 pKa = 6.49DD258 pKa = 4.1GTIDD262 pKa = 3.51QPMGSGPTATHH273 pKa = 6.29LTVSSPNSGRR283 pKa = 11.84YY284 pKa = 8.39FVDD287 pKa = 2.61ITTSEE292 pKa = 4.24GAHH295 pKa = 6.08YY296 pKa = 10.62LDD298 pKa = 6.12DD299 pKa = 4.34ILITFYY305 pKa = 9.84GTPVLDD311 pKa = 3.64RR312 pKa = 11.84VEE314 pKa = 4.45LLNSNLTLDD323 pKa = 3.82PDD325 pKa = 3.89RR326 pKa = 11.84NNIEE330 pKa = 4.05IFVDD334 pKa = 3.46GDD336 pKa = 3.15GDD338 pKa = 3.64YY339 pKa = 10.89EE340 pKa = 4.28YY341 pKa = 10.95AVNGGEE347 pKa = 4.09FQDD350 pKa = 4.47DD351 pKa = 4.26SVFNDD356 pKa = 3.48VPPGINTLVINDD368 pKa = 3.92KK369 pKa = 10.55NGCGTTEE376 pKa = 4.52PIEE379 pKa = 4.07FLVVGYY385 pKa = 9.34PKK387 pKa = 10.75FFTPNNDD394 pKa = 3.4GANDD398 pKa = 3.16TWNIKK403 pKa = 10.53GIEE406 pKa = 4.14TLNNPVVFVFDD417 pKa = 4.47RR418 pKa = 11.84YY419 pKa = 10.88GKK421 pKa = 10.6LLAQLNSNTEE431 pKa = 3.24WDD433 pKa = 3.32GTYY436 pKa = 10.43NGKK439 pKa = 8.84PLPSSDD445 pKa = 2.58YY446 pKa = 8.79WFRR449 pKa = 11.84FEE451 pKa = 4.86YY452 pKa = 10.89EE453 pKa = 3.76EE454 pKa = 4.94DD455 pKa = 3.65EE456 pKa = 4.58EE457 pKa = 5.11GVVVAKK463 pKa = 8.16STRR466 pKa = 11.84ANFTLKK472 pKa = 10.46RR473 pKa = 3.76
MM1 pKa = 7.88RR2 pKa = 11.84FLLALSLVLLFGLQSQAQNSPDD24 pKa = 3.7CRR26 pKa = 11.84SAIPVCADD34 pKa = 3.11APIMGRR40 pKa = 11.84ADD42 pKa = 3.34GGGDD46 pKa = 3.23IDD48 pKa = 5.85DD49 pKa = 5.34FDD51 pKa = 5.93PEE53 pKa = 4.66VITEE57 pKa = 4.4TGCLEE62 pKa = 4.24KK63 pKa = 11.09GSVSNANIEE72 pKa = 4.6NNTSWYY78 pKa = 9.78VFRR81 pKa = 11.84AGTGGQVGFDD91 pKa = 3.54IEE93 pKa = 4.71ALPLPGEE100 pKa = 4.31TTITAEE106 pKa = 3.5WDD108 pKa = 3.61FAVYY112 pKa = 10.33GPDD115 pKa = 3.48VACGEE120 pKa = 4.24ISSGQAQPIRR130 pKa = 11.84CNYY133 pKa = 7.81EE134 pKa = 3.97VNGTRR139 pKa = 11.84YY140 pKa = 8.26TGIGVNPEE148 pKa = 4.11DD149 pKa = 4.1GQAGVPSTTQNLNTYY164 pKa = 9.36DD165 pKa = 3.25EE166 pKa = 4.57WLDD169 pKa = 3.61VLPGEE174 pKa = 4.81VYY176 pKa = 10.98YY177 pKa = 10.27ILINNFSNNTNNDD190 pKa = 3.41PEE192 pKa = 4.34EE193 pKa = 4.78FILTFTGNSVEE204 pKa = 4.38ADD206 pKa = 3.15QNTALDD212 pKa = 3.73CTLRR216 pKa = 11.84DD217 pKa = 3.55EE218 pKa = 5.64FLGLDD223 pKa = 3.36INACEE228 pKa = 4.27GDD230 pKa = 3.67PDD232 pKa = 3.97IEE234 pKa = 4.12LVALNSPAGPDD245 pKa = 3.12IANVAWSVDD254 pKa = 3.85FDD256 pKa = 5.18DD257 pKa = 6.49DD258 pKa = 4.1GTIDD262 pKa = 3.51QPMGSGPTATHH273 pKa = 6.29LTVSSPNSGRR283 pKa = 11.84YY284 pKa = 8.39FVDD287 pKa = 2.61ITTSEE292 pKa = 4.24GAHH295 pKa = 6.08YY296 pKa = 10.62LDD298 pKa = 6.12DD299 pKa = 4.34ILITFYY305 pKa = 9.84GTPVLDD311 pKa = 3.64RR312 pKa = 11.84VEE314 pKa = 4.45LLNSNLTLDD323 pKa = 3.82PDD325 pKa = 3.89RR326 pKa = 11.84NNIEE330 pKa = 4.05IFVDD334 pKa = 3.46GDD336 pKa = 3.15GDD338 pKa = 3.64YY339 pKa = 10.89EE340 pKa = 4.28YY341 pKa = 10.95AVNGGEE347 pKa = 4.09FQDD350 pKa = 4.47DD351 pKa = 4.26SVFNDD356 pKa = 3.48VPPGINTLVINDD368 pKa = 3.92KK369 pKa = 10.55NGCGTTEE376 pKa = 4.52PIEE379 pKa = 4.07FLVVGYY385 pKa = 9.34PKK387 pKa = 10.75FFTPNNDD394 pKa = 3.4GANDD398 pKa = 3.16TWNIKK403 pKa = 10.53GIEE406 pKa = 4.14TLNNPVVFVFDD417 pKa = 4.47RR418 pKa = 11.84YY419 pKa = 10.88GKK421 pKa = 10.6LLAQLNSNTEE431 pKa = 3.24WDD433 pKa = 3.32GTYY436 pKa = 10.43NGKK439 pKa = 8.84PLPSSDD445 pKa = 2.58YY446 pKa = 8.79WFRR449 pKa = 11.84FEE451 pKa = 4.86YY452 pKa = 10.89EE453 pKa = 3.76EE454 pKa = 4.94DD455 pKa = 3.65EE456 pKa = 4.58EE457 pKa = 5.11GVVVAKK463 pKa = 8.16STRR466 pKa = 11.84ANFTLKK472 pKa = 10.46RR473 pKa = 3.76
Molecular weight: 51.57 kDa
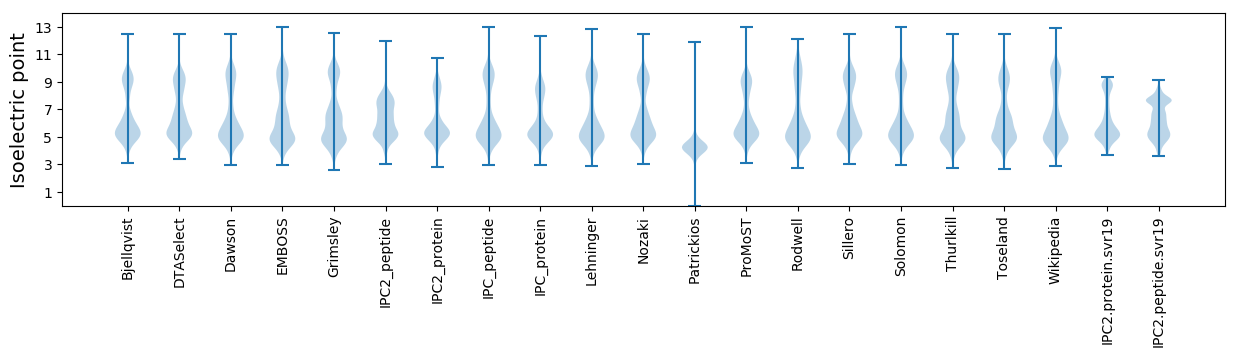
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2L9J9|A0A3G2L9J9_9FLAO Cysteine synthase family protein OS=Euzebyella marina OX=1761453 GN=D1013_16855 PE=4 SV=1
MM1 pKa = 7.26LRR3 pKa = 11.84WTVIFVILAIVAAIFGFGGIAAGAASIAKK32 pKa = 9.38ILFFIFLVLFVISLFMGRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84II53 pKa = 3.67
MM1 pKa = 7.26LRR3 pKa = 11.84WTVIFVILAIVAAIFGFGGIAAGAASIAKK32 pKa = 9.38ILFFIFLVLFVISLFMGRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84II53 pKa = 3.67
Molecular weight: 5.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1409240 |
38 |
5361 |
356.6 |
40.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.486 ± 0.035 | 0.727 ± 0.014 |
5.893 ± 0.053 | 6.981 ± 0.046 |
5.046 ± 0.03 | 6.884 ± 0.047 |
1.858 ± 0.022 | 7.134 ± 0.031 |
7.035 ± 0.074 | 9.421 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.023 | 5.732 ± 0.041 |
3.611 ± 0.024 | 3.479 ± 0.02 |
3.792 ± 0.029 | 6.61 ± 0.033 |
5.51 ± 0.054 | 6.371 ± 0.031 |
1.163 ± 0.016 | 3.96 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
