
Clostridium sp. CAG:793
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
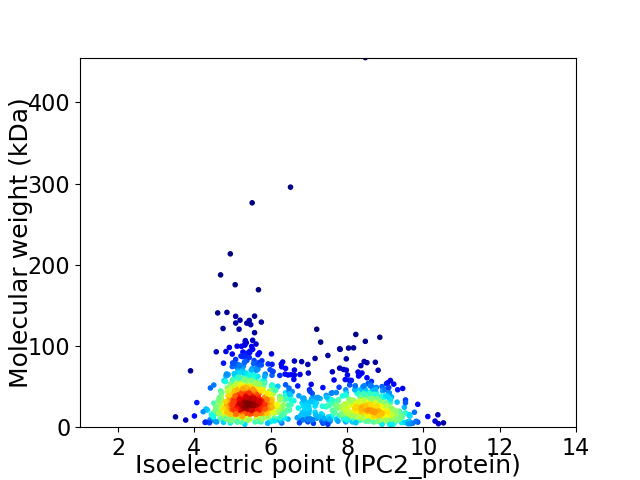
Virtual 2D-PAGE plot for 1163 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
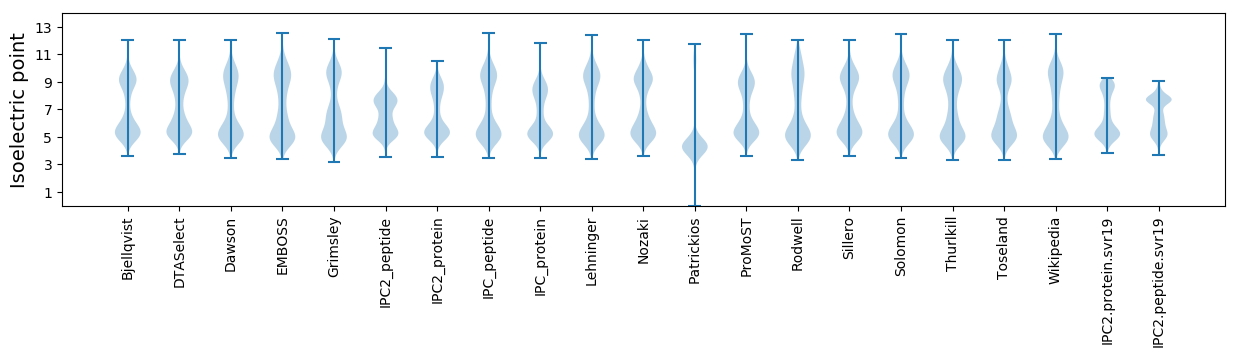
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5NPQ0|R5NPQ0_9CLOT Phosphoesterase RecJ domain protein OS=Clostridium sp. CAG:793 OX=1262840 GN=BN786_00331 PE=4 SV=1
MM1 pKa = 8.32DD2 pKa = 6.03DD3 pKa = 4.98LNNNEE8 pKa = 4.21NNQNNQAGYY17 pKa = 10.23YY18 pKa = 8.66PNNSQDD24 pKa = 3.52PYY26 pKa = 10.31SANSGFGPVNPYY38 pKa = 9.76MNNQDD43 pKa = 2.88GSSFNPYY50 pKa = 9.78GMADD54 pKa = 3.97SNSMPQNPYY63 pKa = 9.83QNSGEE68 pKa = 4.08FNPYY72 pKa = 10.13SLYY75 pKa = 10.9NSTYY79 pKa = 10.03PGAEE83 pKa = 3.87NNQNTVQTSDD93 pKa = 3.15NSHH96 pKa = 7.23DD97 pKa = 3.73SNYY100 pKa = 10.18AQNPNTTVEE109 pKa = 4.1NDD111 pKa = 3.07NGTFSPFDD119 pKa = 3.8LSNEE123 pKa = 3.89SSAVPEE129 pKa = 5.0QSTQMQNNQFNHH141 pKa = 5.31VDD143 pKa = 3.22ASNNQAQTEE152 pKa = 4.11PMQEE156 pKa = 3.59NQMPQEE162 pKa = 4.29PVQEE166 pKa = 4.01NQIPQEE172 pKa = 4.37PIQGNATSYY181 pKa = 11.43DD182 pKa = 3.49PAYY185 pKa = 10.56EE186 pKa = 4.07NMQQQFVQNQGQIRR200 pKa = 11.84NPILSQGLPTDD211 pKa = 4.63DD212 pKa = 4.93GMNNYY217 pKa = 7.87QQNSSFSNNIDD228 pKa = 3.38SYY230 pKa = 10.48NTQPSQDD237 pKa = 3.48YY238 pKa = 10.57SFNGSDD244 pKa = 3.31YY245 pKa = 11.2TMEE248 pKa = 4.45FVKK251 pKa = 10.77AWMGNLYY258 pKa = 10.43EE259 pKa = 4.62KK260 pKa = 10.84AHH262 pKa = 5.77SKK264 pKa = 9.92KK265 pKa = 10.48FNWPAAIFGGIYY277 pKa = 9.59FLYY280 pKa = 10.38RR281 pKa = 11.84KK282 pKa = 9.05MYY284 pKa = 8.73LTGILLIMFSSILTFLMLFLSVKK307 pKa = 10.31VGAGLMALLPVINIVLLLVYY327 pKa = 10.51GFGFYY332 pKa = 10.47PMYY335 pKa = 10.73RR336 pKa = 11.84NFVRR340 pKa = 11.84GKK342 pKa = 9.38LNKK345 pKa = 9.52YY346 pKa = 9.43KK347 pKa = 9.33QTITDD352 pKa = 3.48NSQLVQTASSKK363 pKa = 11.17GGTSPLAVILCCIVVPIISSLVLTLLATVGLFNIFKK399 pKa = 10.39NISGNINTPSNIVSDD414 pKa = 3.71NSIIEE419 pKa = 4.92DD420 pKa = 3.61NTDD423 pKa = 2.98MKK425 pKa = 11.18SYY427 pKa = 9.81TFLNQYY433 pKa = 9.14TIDD436 pKa = 4.02YY437 pKa = 10.74DD438 pKa = 3.76SLTWFFNQSDD448 pKa = 3.91SSLTKK453 pKa = 10.29GDD455 pKa = 3.62YY456 pKa = 10.64KK457 pKa = 10.43LTYY460 pKa = 9.41SGQSIPDD467 pKa = 3.62LKK469 pKa = 11.24SSFNVDD475 pKa = 2.58STTPAGRR482 pKa = 11.84SSLLSTLMSSLQAQAASMNLTVDD505 pKa = 3.25VGTSNFVLGVKK516 pKa = 10.05NYY518 pKa = 9.86YY519 pKa = 10.72AYY521 pKa = 9.9IDD523 pKa = 3.65VASNNTLTRR532 pKa = 11.84YY533 pKa = 9.94YY534 pKa = 10.04IILIPEE540 pKa = 4.52DD541 pKa = 5.38DD542 pKa = 4.07ILFQFTLEE550 pKa = 4.32TNDD553 pKa = 3.3TSIDD557 pKa = 3.55YY558 pKa = 7.66ATNIEE563 pKa = 4.36IVNMLTNISTEE574 pKa = 4.15SSDD577 pKa = 5.1EE578 pKa = 4.02EE579 pKa = 5.81ADD581 pKa = 4.78DD582 pKa = 3.85NTISNDD588 pKa = 3.53TVISNDD594 pKa = 3.06VSDD597 pKa = 4.3ILSNTPVNEE606 pKa = 3.93VTSARR611 pKa = 11.84NTISNTNSLSDD622 pKa = 3.52MLNN625 pKa = 3.08
MM1 pKa = 8.32DD2 pKa = 6.03DD3 pKa = 4.98LNNNEE8 pKa = 4.21NNQNNQAGYY17 pKa = 10.23YY18 pKa = 8.66PNNSQDD24 pKa = 3.52PYY26 pKa = 10.31SANSGFGPVNPYY38 pKa = 9.76MNNQDD43 pKa = 2.88GSSFNPYY50 pKa = 9.78GMADD54 pKa = 3.97SNSMPQNPYY63 pKa = 9.83QNSGEE68 pKa = 4.08FNPYY72 pKa = 10.13SLYY75 pKa = 10.9NSTYY79 pKa = 10.03PGAEE83 pKa = 3.87NNQNTVQTSDD93 pKa = 3.15NSHH96 pKa = 7.23DD97 pKa = 3.73SNYY100 pKa = 10.18AQNPNTTVEE109 pKa = 4.1NDD111 pKa = 3.07NGTFSPFDD119 pKa = 3.8LSNEE123 pKa = 3.89SSAVPEE129 pKa = 5.0QSTQMQNNQFNHH141 pKa = 5.31VDD143 pKa = 3.22ASNNQAQTEE152 pKa = 4.11PMQEE156 pKa = 3.59NQMPQEE162 pKa = 4.29PVQEE166 pKa = 4.01NQIPQEE172 pKa = 4.37PIQGNATSYY181 pKa = 11.43DD182 pKa = 3.49PAYY185 pKa = 10.56EE186 pKa = 4.07NMQQQFVQNQGQIRR200 pKa = 11.84NPILSQGLPTDD211 pKa = 4.63DD212 pKa = 4.93GMNNYY217 pKa = 7.87QQNSSFSNNIDD228 pKa = 3.38SYY230 pKa = 10.48NTQPSQDD237 pKa = 3.48YY238 pKa = 10.57SFNGSDD244 pKa = 3.31YY245 pKa = 11.2TMEE248 pKa = 4.45FVKK251 pKa = 10.77AWMGNLYY258 pKa = 10.43EE259 pKa = 4.62KK260 pKa = 10.84AHH262 pKa = 5.77SKK264 pKa = 9.92KK265 pKa = 10.48FNWPAAIFGGIYY277 pKa = 9.59FLYY280 pKa = 10.38RR281 pKa = 11.84KK282 pKa = 9.05MYY284 pKa = 8.73LTGILLIMFSSILTFLMLFLSVKK307 pKa = 10.31VGAGLMALLPVINIVLLLVYY327 pKa = 10.51GFGFYY332 pKa = 10.47PMYY335 pKa = 10.73RR336 pKa = 11.84NFVRR340 pKa = 11.84GKK342 pKa = 9.38LNKK345 pKa = 9.52YY346 pKa = 9.43KK347 pKa = 9.33QTITDD352 pKa = 3.48NSQLVQTASSKK363 pKa = 11.17GGTSPLAVILCCIVVPIISSLVLTLLATVGLFNIFKK399 pKa = 10.39NISGNINTPSNIVSDD414 pKa = 3.71NSIIEE419 pKa = 4.92DD420 pKa = 3.61NTDD423 pKa = 2.98MKK425 pKa = 11.18SYY427 pKa = 9.81TFLNQYY433 pKa = 9.14TIDD436 pKa = 4.02YY437 pKa = 10.74DD438 pKa = 3.76SLTWFFNQSDD448 pKa = 3.91SSLTKK453 pKa = 10.29GDD455 pKa = 3.62YY456 pKa = 10.64KK457 pKa = 10.43LTYY460 pKa = 9.41SGQSIPDD467 pKa = 3.62LKK469 pKa = 11.24SSFNVDD475 pKa = 2.58STTPAGRR482 pKa = 11.84SSLLSTLMSSLQAQAASMNLTVDD505 pKa = 3.25VGTSNFVLGVKK516 pKa = 10.05NYY518 pKa = 9.86YY519 pKa = 10.72AYY521 pKa = 9.9IDD523 pKa = 3.65VASNNTLTRR532 pKa = 11.84YY533 pKa = 9.94YY534 pKa = 10.04IILIPEE540 pKa = 4.52DD541 pKa = 5.38DD542 pKa = 4.07ILFQFTLEE550 pKa = 4.32TNDD553 pKa = 3.3TSIDD557 pKa = 3.55YY558 pKa = 7.66ATNIEE563 pKa = 4.36IVNMLTNISTEE574 pKa = 4.15SSDD577 pKa = 5.1EE578 pKa = 4.02EE579 pKa = 5.81ADD581 pKa = 4.78DD582 pKa = 3.85NTISNDD588 pKa = 3.53TVISNDD594 pKa = 3.06VSDD597 pKa = 4.3ILSNTPVNEE606 pKa = 3.93VTSARR611 pKa = 11.84NTISNTNSLSDD622 pKa = 3.52MLNN625 pKa = 3.08
Molecular weight: 69.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5NP83|R5NP83_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:793 OX=1262840 GN=BN786_00161 PE=4 SV=1
MM1 pKa = 7.28TFVFSIACAVAAVQILLSLALVFHH25 pKa = 6.67KK26 pKa = 10.52HH27 pKa = 4.73NVLAIVSEE35 pKa = 4.41LATSTGLTLFISRR48 pKa = 11.84YY49 pKa = 8.96LLAWPLPYY57 pKa = 9.87IILLGGAEE65 pKa = 4.56YY66 pKa = 8.86ITVKK70 pKa = 10.53IIILLVIVILKK81 pKa = 8.93LRR83 pKa = 11.84CFTVLTILTHH93 pKa = 5.72PSNRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 9.88SVYY102 pKa = 10.2KK103 pKa = 9.87KK104 pKa = 9.76RR105 pKa = 11.84YY106 pKa = 7.0SGRR109 pKa = 11.84MHH111 pKa = 7.65FISMVYY117 pKa = 9.4RR118 pKa = 11.84SKK120 pKa = 10.78YY121 pKa = 9.62HH122 pKa = 6.35PRR124 pKa = 11.84LKK126 pKa = 10.55LGMPAMVGKK135 pKa = 10.22YY136 pKa = 10.12SNHH139 pKa = 5.95TGIKK143 pKa = 9.08FDD145 pKa = 3.49KK146 pKa = 10.83KK147 pKa = 10.14GFPVFKK153 pKa = 10.16SICTITLPRR162 pKa = 11.84KK163 pKa = 9.46YY164 pKa = 9.88RR165 pKa = 11.84YY166 pKa = 6.9QTRR169 pKa = 11.84DD170 pKa = 3.0VHH172 pKa = 6.41FRR174 pKa = 11.84CASKK178 pKa = 10.74LLYY181 pKa = 10.03KK182 pKa = 10.29RR183 pKa = 11.84AVSNRR188 pKa = 11.84YY189 pKa = 8.48VRR191 pKa = 11.84CKK193 pKa = 8.63FTKK196 pKa = 10.35SEE198 pKa = 4.13LATFKK203 pKa = 11.16CGDD206 pKa = 3.75VPSKK210 pKa = 9.16YY211 pKa = 8.36TWHH214 pKa = 6.52HH215 pKa = 5.59HH216 pKa = 4.36QSAGVLQLVDD226 pKa = 3.91RR227 pKa = 11.84KK228 pKa = 9.29VHH230 pKa = 6.52ANTPHH235 pKa = 6.42IGGYY239 pKa = 8.63SIWGADD245 pKa = 3.21NN246 pKa = 3.73
MM1 pKa = 7.28TFVFSIACAVAAVQILLSLALVFHH25 pKa = 6.67KK26 pKa = 10.52HH27 pKa = 4.73NVLAIVSEE35 pKa = 4.41LATSTGLTLFISRR48 pKa = 11.84YY49 pKa = 8.96LLAWPLPYY57 pKa = 9.87IILLGGAEE65 pKa = 4.56YY66 pKa = 8.86ITVKK70 pKa = 10.53IIILLVIVILKK81 pKa = 8.93LRR83 pKa = 11.84CFTVLTILTHH93 pKa = 5.72PSNRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 9.88SVYY102 pKa = 10.2KK103 pKa = 9.87KK104 pKa = 9.76RR105 pKa = 11.84YY106 pKa = 7.0SGRR109 pKa = 11.84MHH111 pKa = 7.65FISMVYY117 pKa = 9.4RR118 pKa = 11.84SKK120 pKa = 10.78YY121 pKa = 9.62HH122 pKa = 6.35PRR124 pKa = 11.84LKK126 pKa = 10.55LGMPAMVGKK135 pKa = 10.22YY136 pKa = 10.12SNHH139 pKa = 5.95TGIKK143 pKa = 9.08FDD145 pKa = 3.49KK146 pKa = 10.83KK147 pKa = 10.14GFPVFKK153 pKa = 10.16SICTITLPRR162 pKa = 11.84KK163 pKa = 9.46YY164 pKa = 9.88RR165 pKa = 11.84YY166 pKa = 6.9QTRR169 pKa = 11.84DD170 pKa = 3.0VHH172 pKa = 6.41FRR174 pKa = 11.84CASKK178 pKa = 10.74LLYY181 pKa = 10.03KK182 pKa = 10.29RR183 pKa = 11.84AVSNRR188 pKa = 11.84YY189 pKa = 8.48VRR191 pKa = 11.84CKK193 pKa = 8.63FTKK196 pKa = 10.35SEE198 pKa = 4.13LATFKK203 pKa = 11.16CGDD206 pKa = 3.75VPSKK210 pKa = 9.16YY211 pKa = 8.36TWHH214 pKa = 6.52HH215 pKa = 5.59HH216 pKa = 4.36QSAGVLQLVDD226 pKa = 3.91RR227 pKa = 11.84KK228 pKa = 9.29VHH230 pKa = 6.52ANTPHH235 pKa = 6.42IGGYY239 pKa = 8.63SIWGADD245 pKa = 3.21NN246 pKa = 3.73
Molecular weight: 28.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
367022 |
29 |
4295 |
315.6 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.521 ± 0.067 | 1.127 ± 0.024 |
5.797 ± 0.049 | 7.694 ± 0.097 |
3.669 ± 0.062 | 5.793 ± 0.078 |
1.253 ± 0.026 | 10.106 ± 0.12 |
9.575 ± 0.083 | 7.955 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.045 | 6.837 ± 0.078 |
2.657 ± 0.044 | 2.943 ± 0.041 |
3.436 ± 0.058 | 6.2 ± 0.088 |
5.721 ± 0.179 | 6.088 ± 0.06 |
0.638 ± 0.024 | 4.45 ± 0.064 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
