
Torque teno virus 14
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
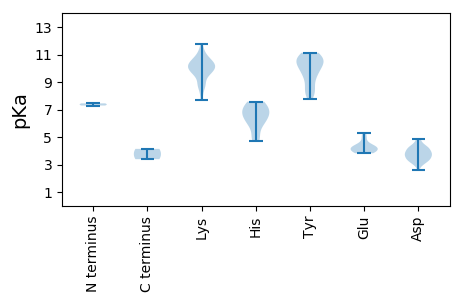
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
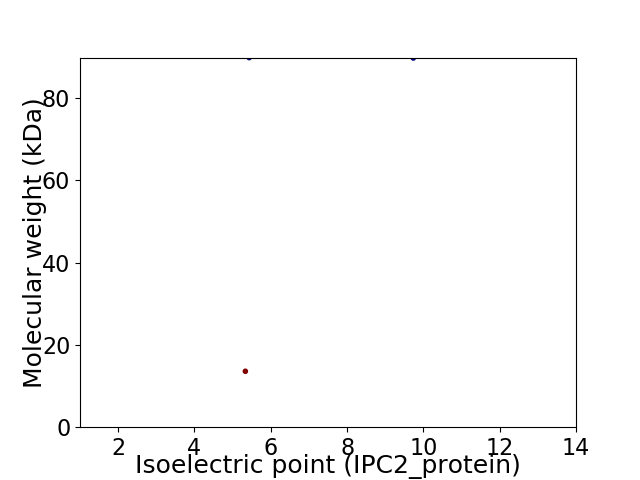
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DUH9|Q9DUH9_9VIRU Uncharacterized protein OS=Torque teno virus 14 OX=687353 PE=4 SV=1
MM1 pKa = 7.29SFWFPPVHH9 pKa = 6.09NVAGQEE15 pKa = 4.23RR16 pKa = 11.84NWFEE20 pKa = 4.2SCFRR24 pKa = 11.84SHH26 pKa = 7.35AAFCGCGNFIGHH38 pKa = 7.57LNNLADD44 pKa = 4.79RR45 pKa = 11.84YY46 pKa = 9.21NHH48 pKa = 6.93PGGPRR53 pKa = 11.84PPGGPGPQAPVPTVRR68 pKa = 11.84ALPALPAAPSRR79 pKa = 11.84PLPPPPPPCRR89 pKa = 11.84GAGGEE94 pKa = 4.24GGGADD99 pKa = 4.27RR100 pKa = 11.84DD101 pKa = 3.77AGGAGDD107 pKa = 4.12AGAVAGRR114 pKa = 11.84GGDD117 pKa = 3.6YY118 pKa = 10.91GPEE121 pKa = 3.87EE122 pKa = 4.36LEE124 pKa = 4.06QPFAAVAGDD133 pKa = 3.62EE134 pKa = 4.14
MM1 pKa = 7.29SFWFPPVHH9 pKa = 6.09NVAGQEE15 pKa = 4.23RR16 pKa = 11.84NWFEE20 pKa = 4.2SCFRR24 pKa = 11.84SHH26 pKa = 7.35AAFCGCGNFIGHH38 pKa = 7.57LNNLADD44 pKa = 4.79RR45 pKa = 11.84YY46 pKa = 9.21NHH48 pKa = 6.93PGGPRR53 pKa = 11.84PPGGPGPQAPVPTVRR68 pKa = 11.84ALPALPAAPSRR79 pKa = 11.84PLPPPPPPCRR89 pKa = 11.84GAGGEE94 pKa = 4.24GGGADD99 pKa = 4.27RR100 pKa = 11.84DD101 pKa = 3.77AGGAGDD107 pKa = 4.12AGAVAGRR114 pKa = 11.84GGDD117 pKa = 3.6YY118 pKa = 10.91GPEE121 pKa = 3.87EE122 pKa = 4.36LEE124 pKa = 4.06QPFAAVAGDD133 pKa = 3.62EE134 pKa = 4.14
Molecular weight: 13.6 kDa
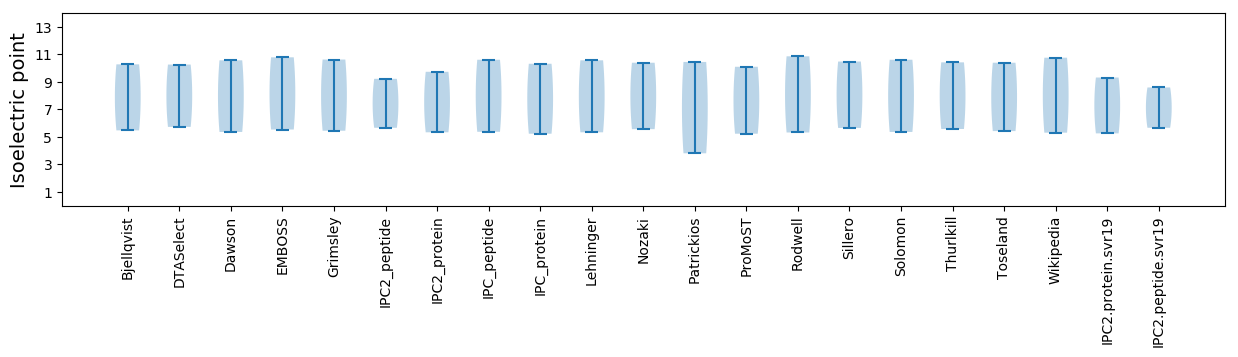
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DUH9|Q9DUH9_9VIRU Uncharacterized protein OS=Torque teno virus 14 OX=687353 PE=4 SV=1
MM1 pKa = 7.45PWGWWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TRR28 pKa = 11.84WGLRR32 pKa = 11.84TRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84AAVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84VRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 9.65HH59 pKa = 4.71YY60 pKa = 9.48RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84FRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 10.18KK74 pKa = 10.16IVLTQWNPQTVRR86 pKa = 11.84KK87 pKa = 9.54CLIRR91 pKa = 11.84GMFPILWCGSGAAGKK106 pKa = 10.19NYY108 pKa = 9.71ALHH111 pKa = 6.54MDD113 pKa = 4.87DD114 pKa = 4.26YY115 pKa = 10.02TPSGGFGGSFSLTTFNLRR133 pKa = 11.84ALYY136 pKa = 9.91DD137 pKa = 3.4EE138 pKa = 4.56HH139 pKa = 6.68MKK141 pKa = 10.56GRR143 pKa = 11.84NRR145 pKa = 11.84WSHH148 pKa = 4.88TNEE151 pKa = 3.92DD152 pKa = 4.38LEE154 pKa = 4.24LALYY158 pKa = 8.34FGCRR162 pKa = 11.84FTFYY166 pKa = 10.65RR167 pKa = 11.84HH168 pKa = 5.84PTTDD172 pKa = 4.47FIVTFSNTPPMMSNQYY188 pKa = 7.77TAPLAHH194 pKa = 7.45PGMLMRR200 pKa = 11.84SKK202 pKa = 11.07YY203 pKa = 10.46KK204 pKa = 10.73LLLPSFKK211 pKa = 10.08TRR213 pKa = 11.84PGGKK217 pKa = 8.0KK218 pKa = 7.72TVSVRR223 pKa = 11.84IRR225 pKa = 11.84PPKK228 pKa = 10.15LFNHH232 pKa = 6.24KK233 pKa = 9.28WYY235 pKa = 10.19AQSDD239 pKa = 4.13LCDD242 pKa = 3.5VNLVQFTVTAADD254 pKa = 3.87FMHH257 pKa = 7.26PFGSPLTEE265 pKa = 4.33TPCVTFQVLGDD276 pKa = 4.67LYY278 pKa = 11.13NKK280 pKa = 9.82CLNIDD285 pKa = 3.88LPKK288 pKa = 10.54VDD290 pKa = 3.74TVKK293 pKa = 10.68NASSPHH299 pKa = 5.17TKK301 pKa = 9.75IQVKK305 pKa = 8.51LTDD308 pKa = 3.52NSGNPTNKK316 pKa = 9.78EE317 pKa = 3.88AVKK320 pKa = 9.52TLYY323 pKa = 10.19EE324 pKa = 3.82QLFSYY329 pKa = 8.76GQYY332 pKa = 8.3WQTFITDD339 pKa = 4.22FMTANPPKK347 pKa = 10.31YY348 pKa = 10.55DD349 pKa = 3.29NGNNKK354 pKa = 9.68KK355 pKa = 10.4LFNDD359 pKa = 3.06NDD361 pKa = 3.78YY362 pKa = 9.48NTFKK366 pKa = 8.77TTFLTKK372 pKa = 9.9KK373 pKa = 10.36DD374 pKa = 3.45SGYY377 pKa = 11.13LFASYY382 pKa = 11.03NPAQVAEE389 pKa = 4.34NIKK392 pKa = 10.55NIRR395 pKa = 11.84DD396 pKa = 3.36QNFALTTGKK405 pKa = 10.19NDD407 pKa = 3.12VFGDD411 pKa = 4.14SKK413 pKa = 11.34AQYY416 pKa = 10.74HH417 pKa = 5.95NATHH421 pKa = 6.92LLDD424 pKa = 4.09YY425 pKa = 10.89KK426 pKa = 10.86LGIYY430 pKa = 10.33SPVFLHH436 pKa = 6.83PGRR439 pKa = 11.84SNIQMWTAYY448 pKa = 10.34RR449 pKa = 11.84DD450 pKa = 3.25IVYY453 pKa = 10.74NPFLDD458 pKa = 3.7KK459 pKa = 11.77GEE461 pKa = 4.33GNKK464 pKa = 8.69VWYY467 pKa = 9.65QYY469 pKa = 8.47HH470 pKa = 6.26TKK472 pKa = 9.83TDD474 pKa = 3.27NKK476 pKa = 10.51YY477 pKa = 11.05DD478 pKa = 3.81PTKK481 pKa = 10.15CRR483 pKa = 11.84YY484 pKa = 8.24MVEE487 pKa = 5.08DD488 pKa = 3.98LPLWSLLHH496 pKa = 6.83GYY498 pKa = 10.26ADD500 pKa = 4.32YY501 pKa = 10.75IISQIKK507 pKa = 9.98YY508 pKa = 9.84GDD510 pKa = 3.73PLVEE514 pKa = 4.0GKK516 pKa = 10.26VLIRR520 pKa = 11.84CPYY523 pKa = 8.33TRR525 pKa = 11.84PALYY529 pKa = 10.34NKK531 pKa = 9.62QSPDD535 pKa = 2.95EE536 pKa = 4.67GYY538 pKa = 10.86VVYY541 pKa = 9.02NTYY544 pKa = 10.87FGMGKK549 pKa = 8.75WVDD552 pKa = 3.4GSGYY556 pKa = 10.08IPIPEE561 pKa = 4.05RR562 pKa = 11.84ARR564 pKa = 11.84WMVMLKK570 pKa = 9.81YY571 pKa = 10.41QLDD574 pKa = 3.87VFHH577 pKa = 7.48DD578 pKa = 4.3LVVCGPWAYY587 pKa = 10.7RR588 pKa = 11.84DD589 pKa = 4.01DD590 pKa = 4.27EE591 pKa = 5.28KK592 pKa = 11.3SVQMPCKK599 pKa = 10.11YY600 pKa = 10.37KK601 pKa = 10.72FRR603 pKa = 11.84FKK605 pKa = 10.35WGGTTLRR612 pKa = 11.84SQVIRR617 pKa = 11.84NPCSGGDD624 pKa = 3.43VAPAYY629 pKa = 8.0PHH631 pKa = 6.67RR632 pKa = 11.84RR633 pKa = 11.84PRR635 pKa = 11.84DD636 pKa = 3.53EE637 pKa = 4.1QVVDD641 pKa = 3.81PRR643 pKa = 11.84VVGPQWSSTPGTRR656 pKa = 11.84DD657 pKa = 3.2EE658 pKa = 4.92DD659 pKa = 3.83CLARR663 pKa = 11.84KK664 pKa = 9.79LSDD667 pKa = 3.05EE668 pKa = 4.41CSRR671 pKa = 11.84SRR673 pKa = 11.84TLAKK677 pKa = 10.39SLLQAQKK684 pKa = 10.89DD685 pKa = 4.04LSCLCQQRR693 pKa = 11.84SPKK696 pKa = 9.82EE697 pKa = 3.83EE698 pKa = 4.17KK699 pKa = 9.01EE700 pKa = 4.01TLLRR704 pKa = 11.84QRR706 pKa = 11.84RR707 pKa = 11.84RR708 pKa = 11.84DD709 pKa = 3.28SPRR712 pKa = 11.84GRR714 pKa = 11.84KK715 pKa = 8.76RR716 pKa = 11.84RR717 pKa = 11.84AARR720 pKa = 11.84RR721 pKa = 11.84RR722 pKa = 11.84TPPRR726 pKa = 11.84KK727 pKa = 8.89SKK729 pKa = 10.3KK730 pKa = 9.98RR731 pKa = 11.84RR732 pKa = 11.84DD733 pKa = 3.62TSSSEE738 pKa = 3.83SSQNSDD744 pKa = 2.62SSGGSWSSWRR754 pKa = 11.84SSSPKK759 pKa = 9.52PSRR762 pKa = 11.84AYY764 pKa = 9.18TT765 pKa = 3.39
MM1 pKa = 7.45PWGWWRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TRR28 pKa = 11.84WGLRR32 pKa = 11.84TRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84AAVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84VRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84YY58 pKa = 9.65HH59 pKa = 4.71YY60 pKa = 9.48RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84FRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 10.18KK74 pKa = 10.16IVLTQWNPQTVRR86 pKa = 11.84KK87 pKa = 9.54CLIRR91 pKa = 11.84GMFPILWCGSGAAGKK106 pKa = 10.19NYY108 pKa = 9.71ALHH111 pKa = 6.54MDD113 pKa = 4.87DD114 pKa = 4.26YY115 pKa = 10.02TPSGGFGGSFSLTTFNLRR133 pKa = 11.84ALYY136 pKa = 9.91DD137 pKa = 3.4EE138 pKa = 4.56HH139 pKa = 6.68MKK141 pKa = 10.56GRR143 pKa = 11.84NRR145 pKa = 11.84WSHH148 pKa = 4.88TNEE151 pKa = 3.92DD152 pKa = 4.38LEE154 pKa = 4.24LALYY158 pKa = 8.34FGCRR162 pKa = 11.84FTFYY166 pKa = 10.65RR167 pKa = 11.84HH168 pKa = 5.84PTTDD172 pKa = 4.47FIVTFSNTPPMMSNQYY188 pKa = 7.77TAPLAHH194 pKa = 7.45PGMLMRR200 pKa = 11.84SKK202 pKa = 11.07YY203 pKa = 10.46KK204 pKa = 10.73LLLPSFKK211 pKa = 10.08TRR213 pKa = 11.84PGGKK217 pKa = 8.0KK218 pKa = 7.72TVSVRR223 pKa = 11.84IRR225 pKa = 11.84PPKK228 pKa = 10.15LFNHH232 pKa = 6.24KK233 pKa = 9.28WYY235 pKa = 10.19AQSDD239 pKa = 4.13LCDD242 pKa = 3.5VNLVQFTVTAADD254 pKa = 3.87FMHH257 pKa = 7.26PFGSPLTEE265 pKa = 4.33TPCVTFQVLGDD276 pKa = 4.67LYY278 pKa = 11.13NKK280 pKa = 9.82CLNIDD285 pKa = 3.88LPKK288 pKa = 10.54VDD290 pKa = 3.74TVKK293 pKa = 10.68NASSPHH299 pKa = 5.17TKK301 pKa = 9.75IQVKK305 pKa = 8.51LTDD308 pKa = 3.52NSGNPTNKK316 pKa = 9.78EE317 pKa = 3.88AVKK320 pKa = 9.52TLYY323 pKa = 10.19EE324 pKa = 3.82QLFSYY329 pKa = 8.76GQYY332 pKa = 8.3WQTFITDD339 pKa = 4.22FMTANPPKK347 pKa = 10.31YY348 pKa = 10.55DD349 pKa = 3.29NGNNKK354 pKa = 9.68KK355 pKa = 10.4LFNDD359 pKa = 3.06NDD361 pKa = 3.78YY362 pKa = 9.48NTFKK366 pKa = 8.77TTFLTKK372 pKa = 9.9KK373 pKa = 10.36DD374 pKa = 3.45SGYY377 pKa = 11.13LFASYY382 pKa = 11.03NPAQVAEE389 pKa = 4.34NIKK392 pKa = 10.55NIRR395 pKa = 11.84DD396 pKa = 3.36QNFALTTGKK405 pKa = 10.19NDD407 pKa = 3.12VFGDD411 pKa = 4.14SKK413 pKa = 11.34AQYY416 pKa = 10.74HH417 pKa = 5.95NATHH421 pKa = 6.92LLDD424 pKa = 4.09YY425 pKa = 10.89KK426 pKa = 10.86LGIYY430 pKa = 10.33SPVFLHH436 pKa = 6.83PGRR439 pKa = 11.84SNIQMWTAYY448 pKa = 10.34RR449 pKa = 11.84DD450 pKa = 3.25IVYY453 pKa = 10.74NPFLDD458 pKa = 3.7KK459 pKa = 11.77GEE461 pKa = 4.33GNKK464 pKa = 8.69VWYY467 pKa = 9.65QYY469 pKa = 8.47HH470 pKa = 6.26TKK472 pKa = 9.83TDD474 pKa = 3.27NKK476 pKa = 10.51YY477 pKa = 11.05DD478 pKa = 3.81PTKK481 pKa = 10.15CRR483 pKa = 11.84YY484 pKa = 8.24MVEE487 pKa = 5.08DD488 pKa = 3.98LPLWSLLHH496 pKa = 6.83GYY498 pKa = 10.26ADD500 pKa = 4.32YY501 pKa = 10.75IISQIKK507 pKa = 9.98YY508 pKa = 9.84GDD510 pKa = 3.73PLVEE514 pKa = 4.0GKK516 pKa = 10.26VLIRR520 pKa = 11.84CPYY523 pKa = 8.33TRR525 pKa = 11.84PALYY529 pKa = 10.34NKK531 pKa = 9.62QSPDD535 pKa = 2.95EE536 pKa = 4.67GYY538 pKa = 10.86VVYY541 pKa = 9.02NTYY544 pKa = 10.87FGMGKK549 pKa = 8.75WVDD552 pKa = 3.4GSGYY556 pKa = 10.08IPIPEE561 pKa = 4.05RR562 pKa = 11.84ARR564 pKa = 11.84WMVMLKK570 pKa = 9.81YY571 pKa = 10.41QLDD574 pKa = 3.87VFHH577 pKa = 7.48DD578 pKa = 4.3LVVCGPWAYY587 pKa = 10.7RR588 pKa = 11.84DD589 pKa = 4.01DD590 pKa = 4.27EE591 pKa = 5.28KK592 pKa = 11.3SVQMPCKK599 pKa = 10.11YY600 pKa = 10.37KK601 pKa = 10.72FRR603 pKa = 11.84FKK605 pKa = 10.35WGGTTLRR612 pKa = 11.84SQVIRR617 pKa = 11.84NPCSGGDD624 pKa = 3.43VAPAYY629 pKa = 8.0PHH631 pKa = 6.67RR632 pKa = 11.84RR633 pKa = 11.84PRR635 pKa = 11.84DD636 pKa = 3.53EE637 pKa = 4.1QVVDD641 pKa = 3.81PRR643 pKa = 11.84VVGPQWSSTPGTRR656 pKa = 11.84DD657 pKa = 3.2EE658 pKa = 4.92DD659 pKa = 3.83CLARR663 pKa = 11.84KK664 pKa = 9.79LSDD667 pKa = 3.05EE668 pKa = 4.41CSRR671 pKa = 11.84SRR673 pKa = 11.84TLAKK677 pKa = 10.39SLLQAQKK684 pKa = 10.89DD685 pKa = 4.04LSCLCQQRR693 pKa = 11.84SPKK696 pKa = 9.82EE697 pKa = 3.83EE698 pKa = 4.17KK699 pKa = 9.01EE700 pKa = 4.01TLLRR704 pKa = 11.84QRR706 pKa = 11.84RR707 pKa = 11.84RR708 pKa = 11.84DD709 pKa = 3.28SPRR712 pKa = 11.84GRR714 pKa = 11.84KK715 pKa = 8.76RR716 pKa = 11.84RR717 pKa = 11.84AARR720 pKa = 11.84RR721 pKa = 11.84RR722 pKa = 11.84TPPRR726 pKa = 11.84KK727 pKa = 8.89SKK729 pKa = 10.3KK730 pKa = 9.98RR731 pKa = 11.84RR732 pKa = 11.84DD733 pKa = 3.62TSSSEE738 pKa = 3.83SSQNSDD744 pKa = 2.62SSGGSWSSWRR754 pKa = 11.84SSSPKK759 pKa = 9.52PSRR762 pKa = 11.84AYY764 pKa = 9.18TT765 pKa = 3.39
Molecular weight: 89.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
899 |
134 |
765 |
449.5 |
51.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.229 ± 4.032 | 2.113 ± 0.442 |
5.562 ± 0.55 | 3.003 ± 1.126 |
4.227 ± 0.506 | 8.12 ± 4.965 |
2.225 ± 0.386 | 2.336 ± 0.806 |
5.895 ± 2.99 | 6.897 ± 1.227 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.891 ± 0.581 | 4.672 ± 0.099 |
7.898 ± 4.321 | 3.448 ± 0.613 |
10.901 ± 2.122 | 6.34 ± 1.702 |
5.784 ± 2.555 | 4.894 ± 0.211 |
2.781 ± 0.653 | 4.783 ± 1.669 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
