
Flavobacterium alvei
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
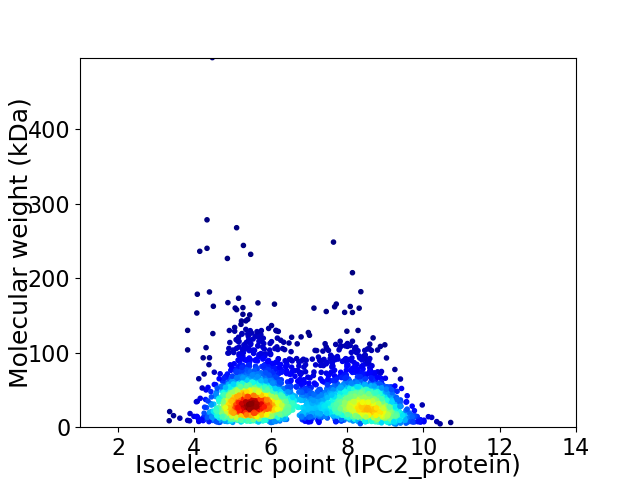
Virtual 2D-PAGE plot for 3153 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5AB32|A0A2S5AB32_9FLAO Uncharacterized protein OS=Flavobacterium alvei OX=2080416 GN=C3L50_10585 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.65KK3 pKa = 9.16MLLKK7 pKa = 10.91KK8 pKa = 10.42NGNAHH13 pKa = 6.56FFNFFLYY20 pKa = 10.46KK21 pKa = 9.37MAFLFLFFILFYY33 pKa = 10.84TKK35 pKa = 11.03GEE37 pKa = 4.1AQNKK41 pKa = 7.68RR42 pKa = 11.84NKK44 pKa = 9.91VEE46 pKa = 4.31LSSSSLTTLSAIKK59 pKa = 10.33KK60 pKa = 5.79EE61 pKa = 4.29TVVAVIGGTGPSDD74 pKa = 4.49DD75 pKa = 4.11FDD77 pKa = 6.82GDD79 pKa = 4.06GVINSVDD86 pKa = 4.32LDD88 pKa = 4.19DD89 pKa = 6.47DD90 pKa = 4.09NDD92 pKa = 5.08GILDD96 pKa = 3.77TDD98 pKa = 3.85EE99 pKa = 4.53TKK101 pKa = 10.97SDD103 pKa = 3.46IPVVSGFDD111 pKa = 3.69PIYY114 pKa = 10.19PVGSTSTTDD123 pKa = 3.2LPLGGRR129 pKa = 11.84AIAYY133 pKa = 7.67NAVSSGGVSYY143 pKa = 10.66HH144 pKa = 7.13CIMEE148 pKa = 4.12ITGKK152 pKa = 10.75GGLPPGIVNIQNNGPLTITGGVPNKK177 pKa = 10.31DD178 pKa = 3.13PYY180 pKa = 9.19VTYY183 pKa = 10.49SVKK186 pKa = 10.18FVPAAGFVLANYY198 pKa = 10.18QNTTPIIGATLTKK211 pKa = 9.54VTVVSADD218 pKa = 3.03VDD220 pKa = 3.7GSSNYY225 pKa = 9.99KK226 pKa = 10.06EE227 pKa = 4.02VTGYY231 pKa = 10.74LPSPNITKK239 pKa = 9.75PVEE242 pKa = 4.07IGTSLSPGGFTQGGGPAAASGYY264 pKa = 10.42VYY266 pKa = 10.74YY267 pKa = 10.85RR268 pKa = 11.84NNDD271 pKa = 3.03GVNPANNAEE280 pKa = 4.05LPPYY284 pKa = 10.53RR285 pKa = 11.84MTIFYY290 pKa = 7.98DD291 pKa = 3.46TFTQADD297 pKa = 4.65LVWGFTGSSSSIRR310 pKa = 11.84SAGSPTMKK318 pKa = 10.24FATIDD323 pKa = 3.26ITDD326 pKa = 3.55ANGNGFSNQFDD337 pKa = 4.54LDD339 pKa = 3.93SDD341 pKa = 4.17GDD343 pKa = 4.01GCSDD347 pKa = 3.83SNEE350 pKa = 4.16AYY352 pKa = 10.47GNSTLADD359 pKa = 3.54TNKK362 pKa = 9.99QYY364 pKa = 11.46GMNAYY369 pKa = 9.06PVGTVAPITPVDD381 pKa = 3.92TFNVTSDD388 pKa = 3.63GKK390 pKa = 10.8VVAASYY396 pKa = 10.42VIVNYY401 pKa = 10.6APVVTITAVSAPVDD415 pKa = 3.66VTNVLIGGTATFSATPNGASASATLKK441 pKa = 9.43WQQKK445 pKa = 10.33LNGSSTWTDD454 pKa = 2.74IVSGTVGGVTYY465 pKa = 10.47AISSFDD471 pKa = 3.46NSLTITGITAALGNSEE487 pKa = 4.1YY488 pKa = 10.69HH489 pKa = 6.69AVLSDD494 pKa = 3.63SGATGYY500 pKa = 8.26FVCGNKK506 pKa = 9.2TSASAKK512 pKa = 10.09LKK514 pKa = 10.04LANISPVANNDD525 pKa = 3.21VLTVNEE531 pKa = 4.36DD532 pKa = 3.03TSGTVNVVTNDD543 pKa = 2.91TDD545 pKa = 4.42ADD547 pKa = 3.95GTLDD551 pKa = 3.57ATTVDD556 pKa = 4.97LDD558 pKa = 3.85PATALIQNTFTVAGQGTYY576 pKa = 8.17TVNNLGVVTFVPVLNFNGSSSISYY600 pKa = 8.12TVNDD604 pKa = 3.44NLGLVSNVATLAVTITSVNDD624 pKa = 3.25APVAKK629 pKa = 10.43NDD631 pKa = 3.34VATTNEE637 pKa = 4.98DD638 pKa = 2.57ISATINVVTNVPGMDD653 pKa = 3.29TDD655 pKa = 3.52IDD657 pKa = 4.06GTVDD661 pKa = 3.16ATKK664 pKa = 10.79VDD666 pKa = 4.98LDD668 pKa = 3.94PATTGIQTTFTVTGQGTYY686 pKa = 8.24TVNNAGLVTFVPVLNYY702 pKa = 10.76NGIASPINYY711 pKa = 8.03TVYY714 pKa = 11.2DD715 pKa = 3.75NLGLVSNISTLTITVVAVNDD735 pKa = 3.71APVAKK740 pKa = 10.28NDD742 pKa = 3.45VVTTNEE748 pKa = 4.18DD749 pKa = 3.05TSITINVVTNALGLDD764 pKa = 3.62TDD766 pKa = 3.53VDD768 pKa = 4.22GTVDD772 pKa = 3.42AATVDD777 pKa = 4.22LDD779 pKa = 4.0LATAGIQKK787 pKa = 8.42TFTITGQGTYY797 pKa = 10.05TIDD800 pKa = 3.24NTGLVTFVPVLNYY813 pKa = 10.53NGSSTINYY821 pKa = 7.3TVNDD825 pKa = 3.62NLGLVSNVTTLTVTITPVNDD845 pKa = 3.2APVAKK850 pKa = 10.43NDD852 pKa = 3.34VATTNEE858 pKa = 4.98DD859 pKa = 2.57ISATINVVTNVPGMDD874 pKa = 3.29TDD876 pKa = 3.52IDD878 pKa = 4.06GTVDD882 pKa = 3.16ATKK885 pKa = 10.79VDD887 pKa = 4.98LDD889 pKa = 3.94PATTGIQTTFTVTGQGTYY907 pKa = 8.86TFNNAGLVTFVPVLNYY923 pKa = 10.76NGIASPINYY932 pKa = 8.03TVYY935 pKa = 11.2DD936 pKa = 3.75NLGLVSNIATLTITVVSVNDD956 pKa = 3.62APVANNDD963 pKa = 3.49TANTEE968 pKa = 3.55KK969 pKa = 9.97DD970 pKa = 3.41TNVIVSVASNDD981 pKa = 3.17TDD983 pKa = 3.31IDD985 pKa = 3.79GTINSATIDD994 pKa = 3.97LDD996 pKa = 3.83LATTGIQTTLTVAGQGTYY1014 pKa = 8.05TANANGTVTFDD1025 pKa = 4.06PLVSFAGTTTPINYY1039 pKa = 9.58IIQDD1043 pKa = 3.63NQAGVSNSATITIIVISDD1061 pKa = 3.39SDD1063 pKa = 3.98GDD1065 pKa = 4.1GVLDD1069 pKa = 4.88KK1070 pKa = 11.24IEE1072 pKa = 4.51IADD1075 pKa = 3.73GTNPNDD1081 pKa = 3.55YY1082 pKa = 10.54CDD1084 pKa = 4.08SKK1086 pKa = 11.49QEE1088 pKa = 4.09NATLEE1093 pKa = 3.97HH1094 pKa = 6.3SAAFLAADD1102 pKa = 4.33CDD1104 pKa = 4.23GDD1106 pKa = 4.36GISNKK1111 pKa = 10.18DD1112 pKa = 3.4EE1113 pKa = 4.14YY1114 pKa = 9.12GTNYY1118 pKa = 9.7GHH1120 pKa = 7.59PFDD1123 pKa = 5.63SNGNGTPDD1131 pKa = 3.27YY1132 pKa = 11.47LEE1134 pKa = 4.93ANNHH1138 pKa = 5.58NLSNSDD1144 pKa = 3.7DD1145 pKa = 3.8DD1146 pKa = 5.89LEE1148 pKa = 4.45IFNGVSPTSDD1158 pKa = 3.2DD1159 pKa = 3.46PKK1161 pKa = 11.21NNVFTIRR1168 pKa = 11.84NIEE1171 pKa = 4.09KK1172 pKa = 10.09YY1173 pKa = 9.8PNNSLHH1179 pKa = 5.91IFNRR1183 pKa = 11.84FGGLVYY1189 pKa = 10.0EE1190 pKa = 4.26AEE1192 pKa = 4.85GYY1194 pKa = 10.48GVGNKK1199 pKa = 8.49FFKK1202 pKa = 10.69GVSEE1206 pKa = 4.17GRR1208 pKa = 11.84STISPSAEE1216 pKa = 4.04LPEE1219 pKa = 4.15GTYY1222 pKa = 10.09FYY1224 pKa = 11.36ALKK1227 pKa = 9.85YY1228 pKa = 9.98VNSAGVTKK1236 pKa = 10.27EE1237 pKa = 3.61RR1238 pKa = 11.84SGYY1241 pKa = 10.77LYY1243 pKa = 10.41INRR1246 pKa = 4.49
MM1 pKa = 7.74KK2 pKa = 10.65KK3 pKa = 9.16MLLKK7 pKa = 10.91KK8 pKa = 10.42NGNAHH13 pKa = 6.56FFNFFLYY20 pKa = 10.46KK21 pKa = 9.37MAFLFLFFILFYY33 pKa = 10.84TKK35 pKa = 11.03GEE37 pKa = 4.1AQNKK41 pKa = 7.68RR42 pKa = 11.84NKK44 pKa = 9.91VEE46 pKa = 4.31LSSSSLTTLSAIKK59 pKa = 10.33KK60 pKa = 5.79EE61 pKa = 4.29TVVAVIGGTGPSDD74 pKa = 4.49DD75 pKa = 4.11FDD77 pKa = 6.82GDD79 pKa = 4.06GVINSVDD86 pKa = 4.32LDD88 pKa = 4.19DD89 pKa = 6.47DD90 pKa = 4.09NDD92 pKa = 5.08GILDD96 pKa = 3.77TDD98 pKa = 3.85EE99 pKa = 4.53TKK101 pKa = 10.97SDD103 pKa = 3.46IPVVSGFDD111 pKa = 3.69PIYY114 pKa = 10.19PVGSTSTTDD123 pKa = 3.2LPLGGRR129 pKa = 11.84AIAYY133 pKa = 7.67NAVSSGGVSYY143 pKa = 10.66HH144 pKa = 7.13CIMEE148 pKa = 4.12ITGKK152 pKa = 10.75GGLPPGIVNIQNNGPLTITGGVPNKK177 pKa = 10.31DD178 pKa = 3.13PYY180 pKa = 9.19VTYY183 pKa = 10.49SVKK186 pKa = 10.18FVPAAGFVLANYY198 pKa = 10.18QNTTPIIGATLTKK211 pKa = 9.54VTVVSADD218 pKa = 3.03VDD220 pKa = 3.7GSSNYY225 pKa = 9.99KK226 pKa = 10.06EE227 pKa = 4.02VTGYY231 pKa = 10.74LPSPNITKK239 pKa = 9.75PVEE242 pKa = 4.07IGTSLSPGGFTQGGGPAAASGYY264 pKa = 10.42VYY266 pKa = 10.74YY267 pKa = 10.85RR268 pKa = 11.84NNDD271 pKa = 3.03GVNPANNAEE280 pKa = 4.05LPPYY284 pKa = 10.53RR285 pKa = 11.84MTIFYY290 pKa = 7.98DD291 pKa = 3.46TFTQADD297 pKa = 4.65LVWGFTGSSSSIRR310 pKa = 11.84SAGSPTMKK318 pKa = 10.24FATIDD323 pKa = 3.26ITDD326 pKa = 3.55ANGNGFSNQFDD337 pKa = 4.54LDD339 pKa = 3.93SDD341 pKa = 4.17GDD343 pKa = 4.01GCSDD347 pKa = 3.83SNEE350 pKa = 4.16AYY352 pKa = 10.47GNSTLADD359 pKa = 3.54TNKK362 pKa = 9.99QYY364 pKa = 11.46GMNAYY369 pKa = 9.06PVGTVAPITPVDD381 pKa = 3.92TFNVTSDD388 pKa = 3.63GKK390 pKa = 10.8VVAASYY396 pKa = 10.42VIVNYY401 pKa = 10.6APVVTITAVSAPVDD415 pKa = 3.66VTNVLIGGTATFSATPNGASASATLKK441 pKa = 9.43WQQKK445 pKa = 10.33LNGSSTWTDD454 pKa = 2.74IVSGTVGGVTYY465 pKa = 10.47AISSFDD471 pKa = 3.46NSLTITGITAALGNSEE487 pKa = 4.1YY488 pKa = 10.69HH489 pKa = 6.69AVLSDD494 pKa = 3.63SGATGYY500 pKa = 8.26FVCGNKK506 pKa = 9.2TSASAKK512 pKa = 10.09LKK514 pKa = 10.04LANISPVANNDD525 pKa = 3.21VLTVNEE531 pKa = 4.36DD532 pKa = 3.03TSGTVNVVTNDD543 pKa = 2.91TDD545 pKa = 4.42ADD547 pKa = 3.95GTLDD551 pKa = 3.57ATTVDD556 pKa = 4.97LDD558 pKa = 3.85PATALIQNTFTVAGQGTYY576 pKa = 8.17TVNNLGVVTFVPVLNFNGSSSISYY600 pKa = 8.12TVNDD604 pKa = 3.44NLGLVSNVATLAVTITSVNDD624 pKa = 3.25APVAKK629 pKa = 10.43NDD631 pKa = 3.34VATTNEE637 pKa = 4.98DD638 pKa = 2.57ISATINVVTNVPGMDD653 pKa = 3.29TDD655 pKa = 3.52IDD657 pKa = 4.06GTVDD661 pKa = 3.16ATKK664 pKa = 10.79VDD666 pKa = 4.98LDD668 pKa = 3.94PATTGIQTTFTVTGQGTYY686 pKa = 8.24TVNNAGLVTFVPVLNYY702 pKa = 10.76NGIASPINYY711 pKa = 8.03TVYY714 pKa = 11.2DD715 pKa = 3.75NLGLVSNISTLTITVVAVNDD735 pKa = 3.71APVAKK740 pKa = 10.28NDD742 pKa = 3.45VVTTNEE748 pKa = 4.18DD749 pKa = 3.05TSITINVVTNALGLDD764 pKa = 3.62TDD766 pKa = 3.53VDD768 pKa = 4.22GTVDD772 pKa = 3.42AATVDD777 pKa = 4.22LDD779 pKa = 4.0LATAGIQKK787 pKa = 8.42TFTITGQGTYY797 pKa = 10.05TIDD800 pKa = 3.24NTGLVTFVPVLNYY813 pKa = 10.53NGSSTINYY821 pKa = 7.3TVNDD825 pKa = 3.62NLGLVSNVTTLTVTITPVNDD845 pKa = 3.2APVAKK850 pKa = 10.43NDD852 pKa = 3.34VATTNEE858 pKa = 4.98DD859 pKa = 2.57ISATINVVTNVPGMDD874 pKa = 3.29TDD876 pKa = 3.52IDD878 pKa = 4.06GTVDD882 pKa = 3.16ATKK885 pKa = 10.79VDD887 pKa = 4.98LDD889 pKa = 3.94PATTGIQTTFTVTGQGTYY907 pKa = 8.86TFNNAGLVTFVPVLNYY923 pKa = 10.76NGIASPINYY932 pKa = 8.03TVYY935 pKa = 11.2DD936 pKa = 3.75NLGLVSNIATLTITVVSVNDD956 pKa = 3.62APVANNDD963 pKa = 3.49TANTEE968 pKa = 3.55KK969 pKa = 9.97DD970 pKa = 3.41TNVIVSVASNDD981 pKa = 3.17TDD983 pKa = 3.31IDD985 pKa = 3.79GTINSATIDD994 pKa = 3.97LDD996 pKa = 3.83LATTGIQTTLTVAGQGTYY1014 pKa = 8.05TANANGTVTFDD1025 pKa = 4.06PLVSFAGTTTPINYY1039 pKa = 9.58IIQDD1043 pKa = 3.63NQAGVSNSATITIIVISDD1061 pKa = 3.39SDD1063 pKa = 3.98GDD1065 pKa = 4.1GVLDD1069 pKa = 4.88KK1070 pKa = 11.24IEE1072 pKa = 4.51IADD1075 pKa = 3.73GTNPNDD1081 pKa = 3.55YY1082 pKa = 10.54CDD1084 pKa = 4.08SKK1086 pKa = 11.49QEE1088 pKa = 4.09NATLEE1093 pKa = 3.97HH1094 pKa = 6.3SAAFLAADD1102 pKa = 4.33CDD1104 pKa = 4.23GDD1106 pKa = 4.36GISNKK1111 pKa = 10.18DD1112 pKa = 3.4EE1113 pKa = 4.14YY1114 pKa = 9.12GTNYY1118 pKa = 9.7GHH1120 pKa = 7.59PFDD1123 pKa = 5.63SNGNGTPDD1131 pKa = 3.27YY1132 pKa = 11.47LEE1134 pKa = 4.93ANNHH1138 pKa = 5.58NLSNSDD1144 pKa = 3.7DD1145 pKa = 3.8DD1146 pKa = 5.89LEE1148 pKa = 4.45IFNGVSPTSDD1158 pKa = 3.2DD1159 pKa = 3.46PKK1161 pKa = 11.21NNVFTIRR1168 pKa = 11.84NIEE1171 pKa = 4.09KK1172 pKa = 10.09YY1173 pKa = 9.8PNNSLHH1179 pKa = 5.91IFNRR1183 pKa = 11.84FGGLVYY1189 pKa = 10.0EE1190 pKa = 4.26AEE1192 pKa = 4.85GYY1194 pKa = 10.48GVGNKK1199 pKa = 8.49FFKK1202 pKa = 10.69GVSEE1206 pKa = 4.17GRR1208 pKa = 11.84STISPSAEE1216 pKa = 4.04LPEE1219 pKa = 4.15GTYY1222 pKa = 10.09FYY1224 pKa = 11.36ALKK1227 pKa = 9.85YY1228 pKa = 9.98VNSAGVTKK1236 pKa = 10.27EE1237 pKa = 3.61RR1238 pKa = 11.84SGYY1241 pKa = 10.77LYY1243 pKa = 10.41INRR1246 pKa = 4.49
Molecular weight: 130.02 kDa
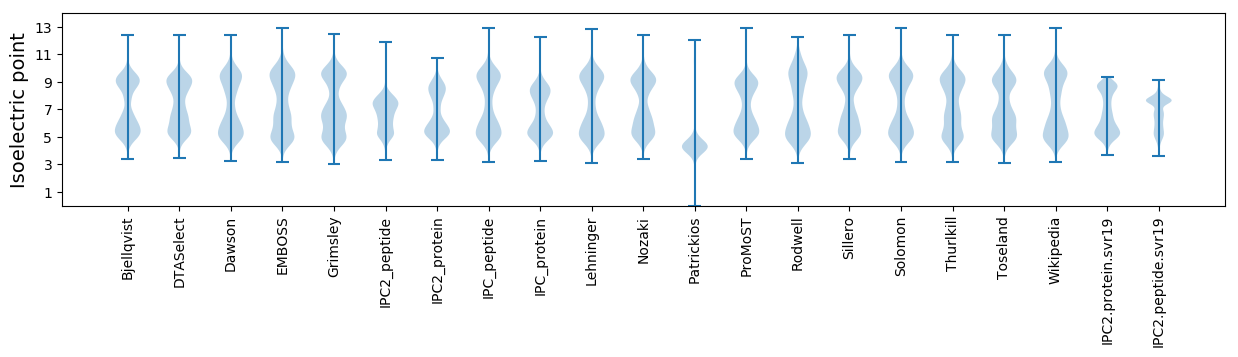
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5A1R5|A0A2S5A1R5_9FLAO YceI domain-containing protein OS=Flavobacterium alvei OX=2080416 GN=C3L50_15435 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1138329 |
36 |
4741 |
361.0 |
40.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.443 ± 0.038 | 0.802 ± 0.014 |
5.247 ± 0.03 | 6.196 ± 0.049 |
5.353 ± 0.04 | 6.459 ± 0.044 |
1.576 ± 0.021 | 8.175 ± 0.042 |
8.086 ± 0.054 | 9.002 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.203 ± 0.026 | 6.527 ± 0.047 |
3.476 ± 0.025 | 3.387 ± 0.026 |
3.004 ± 0.028 | 6.63 ± 0.042 |
6.063 ± 0.069 | 6.193 ± 0.032 |
1.12 ± 0.018 | 4.058 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
