
Aspergillus fumigatus polymycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
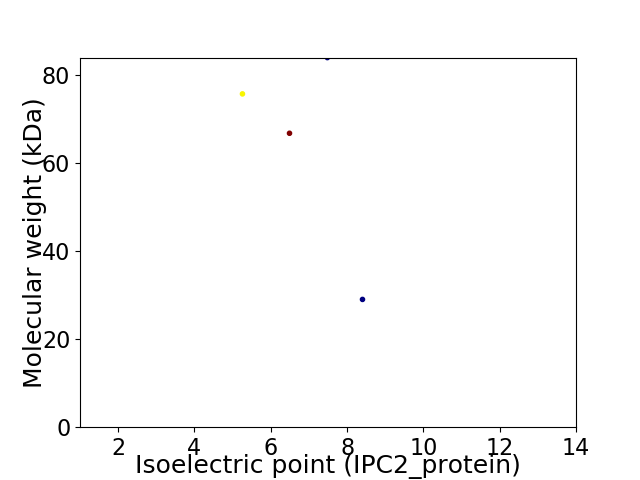
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344X3Q4|A0A344X3Q4_9VIRU Methyl transferase OS=Aspergillus fumigatus polymycovirus 1 OX=2250469 PE=4 SV=1
MM1 pKa = 7.93ADD3 pKa = 3.53LTRR6 pKa = 11.84LRR8 pKa = 11.84NIVLSGDD15 pKa = 3.5MKK17 pKa = 10.96VALTAAILYY26 pKa = 8.95HH27 pKa = 6.68LSPNPSAITDD37 pKa = 3.62YY38 pKa = 11.39GVDD41 pKa = 3.46EE42 pKa = 4.7VKK44 pKa = 10.81SVVSWLVSSPGMFYY58 pKa = 11.25GDD60 pKa = 3.27VVAATGITPDD70 pKa = 3.47ILSTFLPALDD80 pKa = 4.73DD81 pKa = 3.81QACADD86 pKa = 3.88AASLANAMATPDD98 pKa = 3.89AEE100 pKa = 4.09WDD102 pKa = 3.55RR103 pKa = 11.84EE104 pKa = 3.94HH105 pKa = 7.63RR106 pKa = 11.84NRR108 pKa = 11.84FTSALAHH115 pKa = 6.41HH116 pKa = 6.59SADD119 pKa = 3.55AARR122 pKa = 11.84LAAQSSAEE130 pKa = 4.03FAAVDD135 pKa = 3.44AMEE138 pKa = 4.19EE139 pKa = 3.78AGYY142 pKa = 10.64EE143 pKa = 4.24NQDD146 pKa = 3.33HH147 pKa = 6.9ARR149 pKa = 11.84TAVNEE154 pKa = 3.91LNRR157 pKa = 11.84VYY159 pKa = 11.09GNTYY163 pKa = 10.04RR164 pKa = 11.84VFTRR168 pKa = 11.84QHH170 pKa = 4.69ARR172 pKa = 11.84SRR174 pKa = 11.84VAIAASFKK182 pKa = 10.37IDD184 pKa = 3.28DD185 pKa = 4.57HH186 pKa = 6.05GTLFHH191 pKa = 7.12VLTPWCLSRR200 pKa = 11.84TSAMIYY206 pKa = 8.32VAVIARR212 pKa = 11.84HH213 pKa = 4.01VRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84HH218 pKa = 4.78IEE220 pKa = 3.72EE221 pKa = 3.8RR222 pKa = 11.84ASRR225 pKa = 11.84LARR228 pKa = 11.84TALTYY233 pKa = 10.36AARR236 pKa = 11.84AGLEE240 pKa = 3.84AVRR243 pKa = 11.84AIIEE247 pKa = 3.85PAIAIVTSFRR257 pKa = 11.84YY258 pKa = 10.2DD259 pKa = 3.27PAKK262 pKa = 10.33IAFVDD267 pKa = 3.45DD268 pKa = 4.19FDD270 pKa = 4.74RR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.3SITTRR279 pKa = 11.84SPPTLVAFACVHH291 pKa = 5.0MCGNQDD297 pKa = 3.66VVSLAARR304 pKa = 11.84LRR306 pKa = 11.84MEE308 pKa = 4.94SLPQLHH314 pKa = 6.03EE315 pKa = 4.11TPPAVVEE322 pKa = 4.32YY323 pKa = 9.52TDD325 pKa = 4.87HH326 pKa = 8.15VDD328 pKa = 4.26DD329 pKa = 4.68EE330 pKa = 4.58QSALLMFSRR339 pKa = 11.84VLKK342 pKa = 10.29RR343 pKa = 11.84LRR345 pKa = 11.84HH346 pKa = 5.08EE347 pKa = 4.55RR348 pKa = 11.84DD349 pKa = 3.2TGVHH353 pKa = 6.36GPRR356 pKa = 11.84LNKK359 pKa = 10.48DD360 pKa = 3.01PMLGYY365 pKa = 9.7IARR368 pKa = 11.84GVRR371 pKa = 11.84RR372 pKa = 11.84SEE374 pKa = 4.22RR375 pKa = 11.84NTMACINRR383 pKa = 11.84VEE385 pKa = 4.19EE386 pKa = 4.08AVSFLRR392 pKa = 11.84SKK394 pKa = 11.16GVDD397 pKa = 3.18TLNTLFVIEE406 pKa = 4.4WGGEE410 pKa = 3.63FDD412 pKa = 4.36YY413 pKa = 9.22PTVMAALAVARR424 pKa = 11.84LDD426 pKa = 3.48VCVDD430 pKa = 3.7IGPSGIDD437 pKa = 4.46LPGADD442 pKa = 4.88AKK444 pKa = 11.41AEE446 pKa = 4.54DD447 pKa = 3.9GTEE450 pKa = 4.02EE451 pKa = 4.13YY452 pKa = 10.7NYY454 pKa = 11.34SLLLASAEE462 pKa = 4.11EE463 pKa = 4.1RR464 pKa = 11.84RR465 pKa = 11.84MPRR468 pKa = 11.84MPRR471 pKa = 11.84VPYY474 pKa = 10.32PSDD477 pKa = 3.37EE478 pKa = 4.09PLEE481 pKa = 4.39AKK483 pKa = 9.9LRR485 pKa = 11.84RR486 pKa = 11.84VAQFYY491 pKa = 10.85SLSGSANIAYY501 pKa = 9.59ISGGVTQNEE510 pKa = 4.22GVPVNICIDD519 pKa = 3.42TANRR523 pKa = 11.84LNAIRR528 pKa = 11.84NVTPDD533 pKa = 3.21VNVVYY538 pKa = 10.74ACAEE542 pKa = 3.88ALLPPLCGHH551 pKa = 6.21GLEE554 pKa = 5.3LGPDD558 pKa = 4.15DD559 pKa = 4.86YY560 pKa = 11.36LQCASLVDD568 pKa = 4.4EE569 pKa = 5.57DD570 pKa = 5.53CPACNAHH577 pKa = 5.81YY578 pKa = 10.11RR579 pKa = 11.84ATAVLSAACADD590 pKa = 3.86RR591 pKa = 11.84QVRR594 pKa = 11.84IVKK597 pKa = 9.38PRR599 pKa = 11.84SAYY602 pKa = 9.64AHH604 pKa = 6.42NGHH607 pKa = 7.18LSLEE611 pKa = 4.41LVPGHH616 pKa = 6.04PTCVDD621 pKa = 3.39DD622 pKa = 4.71TLLTVDD628 pKa = 4.4SMVAANVLRR637 pKa = 11.84NHH639 pKa = 7.01DD640 pKa = 3.84WGDD643 pKa = 3.69EE644 pKa = 4.1PVTPPAGGNPTSPTLARR661 pKa = 11.84VTAEE665 pKa = 3.69NMRR668 pKa = 11.84TVYY671 pKa = 10.58SALNNGAVGPFDD683 pKa = 4.64YY684 pKa = 11.32DD685 pKa = 3.72DD686 pKa = 3.99MASIAASIAPP696 pKa = 3.85
MM1 pKa = 7.93ADD3 pKa = 3.53LTRR6 pKa = 11.84LRR8 pKa = 11.84NIVLSGDD15 pKa = 3.5MKK17 pKa = 10.96VALTAAILYY26 pKa = 8.95HH27 pKa = 6.68LSPNPSAITDD37 pKa = 3.62YY38 pKa = 11.39GVDD41 pKa = 3.46EE42 pKa = 4.7VKK44 pKa = 10.81SVVSWLVSSPGMFYY58 pKa = 11.25GDD60 pKa = 3.27VVAATGITPDD70 pKa = 3.47ILSTFLPALDD80 pKa = 4.73DD81 pKa = 3.81QACADD86 pKa = 3.88AASLANAMATPDD98 pKa = 3.89AEE100 pKa = 4.09WDD102 pKa = 3.55RR103 pKa = 11.84EE104 pKa = 3.94HH105 pKa = 7.63RR106 pKa = 11.84NRR108 pKa = 11.84FTSALAHH115 pKa = 6.41HH116 pKa = 6.59SADD119 pKa = 3.55AARR122 pKa = 11.84LAAQSSAEE130 pKa = 4.03FAAVDD135 pKa = 3.44AMEE138 pKa = 4.19EE139 pKa = 3.78AGYY142 pKa = 10.64EE143 pKa = 4.24NQDD146 pKa = 3.33HH147 pKa = 6.9ARR149 pKa = 11.84TAVNEE154 pKa = 3.91LNRR157 pKa = 11.84VYY159 pKa = 11.09GNTYY163 pKa = 10.04RR164 pKa = 11.84VFTRR168 pKa = 11.84QHH170 pKa = 4.69ARR172 pKa = 11.84SRR174 pKa = 11.84VAIAASFKK182 pKa = 10.37IDD184 pKa = 3.28DD185 pKa = 4.57HH186 pKa = 6.05GTLFHH191 pKa = 7.12VLTPWCLSRR200 pKa = 11.84TSAMIYY206 pKa = 8.32VAVIARR212 pKa = 11.84HH213 pKa = 4.01VRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84HH218 pKa = 4.78IEE220 pKa = 3.72EE221 pKa = 3.8RR222 pKa = 11.84ASRR225 pKa = 11.84LARR228 pKa = 11.84TALTYY233 pKa = 10.36AARR236 pKa = 11.84AGLEE240 pKa = 3.84AVRR243 pKa = 11.84AIIEE247 pKa = 3.85PAIAIVTSFRR257 pKa = 11.84YY258 pKa = 10.2DD259 pKa = 3.27PAKK262 pKa = 10.33IAFVDD267 pKa = 3.45DD268 pKa = 4.19FDD270 pKa = 4.74RR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.3SITTRR279 pKa = 11.84SPPTLVAFACVHH291 pKa = 5.0MCGNQDD297 pKa = 3.66VVSLAARR304 pKa = 11.84LRR306 pKa = 11.84MEE308 pKa = 4.94SLPQLHH314 pKa = 6.03EE315 pKa = 4.11TPPAVVEE322 pKa = 4.32YY323 pKa = 9.52TDD325 pKa = 4.87HH326 pKa = 8.15VDD328 pKa = 4.26DD329 pKa = 4.68EE330 pKa = 4.58QSALLMFSRR339 pKa = 11.84VLKK342 pKa = 10.29RR343 pKa = 11.84LRR345 pKa = 11.84HH346 pKa = 5.08EE347 pKa = 4.55RR348 pKa = 11.84DD349 pKa = 3.2TGVHH353 pKa = 6.36GPRR356 pKa = 11.84LNKK359 pKa = 10.48DD360 pKa = 3.01PMLGYY365 pKa = 9.7IARR368 pKa = 11.84GVRR371 pKa = 11.84RR372 pKa = 11.84SEE374 pKa = 4.22RR375 pKa = 11.84NTMACINRR383 pKa = 11.84VEE385 pKa = 4.19EE386 pKa = 4.08AVSFLRR392 pKa = 11.84SKK394 pKa = 11.16GVDD397 pKa = 3.18TLNTLFVIEE406 pKa = 4.4WGGEE410 pKa = 3.63FDD412 pKa = 4.36YY413 pKa = 9.22PTVMAALAVARR424 pKa = 11.84LDD426 pKa = 3.48VCVDD430 pKa = 3.7IGPSGIDD437 pKa = 4.46LPGADD442 pKa = 4.88AKK444 pKa = 11.41AEE446 pKa = 4.54DD447 pKa = 3.9GTEE450 pKa = 4.02EE451 pKa = 4.13YY452 pKa = 10.7NYY454 pKa = 11.34SLLLASAEE462 pKa = 4.11EE463 pKa = 4.1RR464 pKa = 11.84RR465 pKa = 11.84MPRR468 pKa = 11.84MPRR471 pKa = 11.84VPYY474 pKa = 10.32PSDD477 pKa = 3.37EE478 pKa = 4.09PLEE481 pKa = 4.39AKK483 pKa = 9.9LRR485 pKa = 11.84RR486 pKa = 11.84VAQFYY491 pKa = 10.85SLSGSANIAYY501 pKa = 9.59ISGGVTQNEE510 pKa = 4.22GVPVNICIDD519 pKa = 3.42TANRR523 pKa = 11.84LNAIRR528 pKa = 11.84NVTPDD533 pKa = 3.21VNVVYY538 pKa = 10.74ACAEE542 pKa = 3.88ALLPPLCGHH551 pKa = 6.21GLEE554 pKa = 5.3LGPDD558 pKa = 4.15DD559 pKa = 4.86YY560 pKa = 11.36LQCASLVDD568 pKa = 4.4EE569 pKa = 5.57DD570 pKa = 5.53CPACNAHH577 pKa = 5.81YY578 pKa = 10.11RR579 pKa = 11.84ATAVLSAACADD590 pKa = 3.86RR591 pKa = 11.84QVRR594 pKa = 11.84IVKK597 pKa = 9.38PRR599 pKa = 11.84SAYY602 pKa = 9.64AHH604 pKa = 6.42NGHH607 pKa = 7.18LSLEE611 pKa = 4.41LVPGHH616 pKa = 6.04PTCVDD621 pKa = 3.39DD622 pKa = 4.71TLLTVDD628 pKa = 4.4SMVAANVLRR637 pKa = 11.84NHH639 pKa = 7.01DD640 pKa = 3.84WGDD643 pKa = 3.69EE644 pKa = 4.1PVTPPAGGNPTSPTLARR661 pKa = 11.84VTAEE665 pKa = 3.69NMRR668 pKa = 11.84TVYY671 pKa = 10.58SALNNGAVGPFDD683 pKa = 4.64YY684 pKa = 11.32DD685 pKa = 3.72DD686 pKa = 3.99MASIAASIAPP696 pKa = 3.85
Molecular weight: 75.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344X3Q5|A0A344X3Q5_9VIRU PAS-rp OS=Aspergillus fumigatus polymycovirus 1 OX=2250469 PE=4 SV=1
MM1 pKa = 7.49SPAPILSAEE10 pKa = 4.21QAAHH14 pKa = 7.15LAQLTVDD21 pKa = 4.28DD22 pKa = 4.29VSLVIKK28 pKa = 10.25LASLGLKK35 pKa = 10.34AKK37 pKa = 10.34DD38 pKa = 3.26IQQYY42 pKa = 9.52AAEE45 pKa = 4.24IAEE48 pKa = 4.66GGDD51 pKa = 3.69PEE53 pKa = 5.53IDD55 pKa = 3.66LAPGAPRR62 pKa = 11.84PVVIHH67 pKa = 6.35AWSFVKK73 pKa = 10.56DD74 pKa = 3.22RR75 pKa = 11.84GQYY78 pKa = 9.4HH79 pKa = 5.04STYY82 pKa = 10.41GLSSAQAGEE91 pKa = 4.35LLDD94 pKa = 5.6LLHH97 pKa = 7.12TDD99 pKa = 3.57PKK101 pKa = 10.96DD102 pKa = 3.34AVKK105 pKa = 10.53RR106 pKa = 11.84IQALVSEE113 pKa = 4.92RR114 pKa = 11.84LRR116 pKa = 11.84LRR118 pKa = 11.84GSSRR122 pKa = 11.84PVVVDD127 pKa = 3.86LEE129 pKa = 4.61GAPSSTPAKK138 pKa = 10.26GGAPPSGDD146 pKa = 3.0GGLSLLRR153 pKa = 11.84QEE155 pKa = 4.75IKK157 pKa = 10.56RR158 pKa = 11.84NSGVYY163 pKa = 10.16GSYY166 pKa = 10.46QFTAYY171 pKa = 7.23PTNRR175 pKa = 11.84PGGQAFAVGLGGGLYY190 pKa = 10.72AFGQSKK196 pKa = 10.19AVAVAAARR204 pKa = 11.84IARR207 pKa = 11.84VKK209 pKa = 10.56GRR211 pKa = 11.84SDD213 pKa = 3.61DD214 pKa = 3.55LVRR217 pKa = 11.84PYY219 pKa = 10.62IHH221 pKa = 6.65FWVEE225 pKa = 3.93GSSPRR230 pKa = 11.84AASDD234 pKa = 3.15IPAKK238 pKa = 10.57VWDD241 pKa = 4.34GRR243 pKa = 11.84LGPHH247 pKa = 5.99TEE249 pKa = 4.34PPAEE253 pKa = 4.33PATKK257 pKa = 10.28AKK259 pKa = 10.11TEE261 pKa = 4.08GPTTSGGRR269 pKa = 11.84ASTPKK274 pKa = 10.8GGDD277 pKa = 3.07AAA279 pKa = 4.65
MM1 pKa = 7.49SPAPILSAEE10 pKa = 4.21QAAHH14 pKa = 7.15LAQLTVDD21 pKa = 4.28DD22 pKa = 4.29VSLVIKK28 pKa = 10.25LASLGLKK35 pKa = 10.34AKK37 pKa = 10.34DD38 pKa = 3.26IQQYY42 pKa = 9.52AAEE45 pKa = 4.24IAEE48 pKa = 4.66GGDD51 pKa = 3.69PEE53 pKa = 5.53IDD55 pKa = 3.66LAPGAPRR62 pKa = 11.84PVVIHH67 pKa = 6.35AWSFVKK73 pKa = 10.56DD74 pKa = 3.22RR75 pKa = 11.84GQYY78 pKa = 9.4HH79 pKa = 5.04STYY82 pKa = 10.41GLSSAQAGEE91 pKa = 4.35LLDD94 pKa = 5.6LLHH97 pKa = 7.12TDD99 pKa = 3.57PKK101 pKa = 10.96DD102 pKa = 3.34AVKK105 pKa = 10.53RR106 pKa = 11.84IQALVSEE113 pKa = 4.92RR114 pKa = 11.84LRR116 pKa = 11.84LRR118 pKa = 11.84GSSRR122 pKa = 11.84PVVVDD127 pKa = 3.86LEE129 pKa = 4.61GAPSSTPAKK138 pKa = 10.26GGAPPSGDD146 pKa = 3.0GGLSLLRR153 pKa = 11.84QEE155 pKa = 4.75IKK157 pKa = 10.56RR158 pKa = 11.84NSGVYY163 pKa = 10.16GSYY166 pKa = 10.46QFTAYY171 pKa = 7.23PTNRR175 pKa = 11.84PGGQAFAVGLGGGLYY190 pKa = 10.72AFGQSKK196 pKa = 10.19AVAVAAARR204 pKa = 11.84IARR207 pKa = 11.84VKK209 pKa = 10.56GRR211 pKa = 11.84SDD213 pKa = 3.61DD214 pKa = 3.55LVRR217 pKa = 11.84PYY219 pKa = 10.62IHH221 pKa = 6.65FWVEE225 pKa = 3.93GSSPRR230 pKa = 11.84AASDD234 pKa = 3.15IPAKK238 pKa = 10.57VWDD241 pKa = 4.34GRR243 pKa = 11.84LGPHH247 pKa = 5.99TEE249 pKa = 4.34PPAEE253 pKa = 4.33PATKK257 pKa = 10.28AKK259 pKa = 10.11TEE261 pKa = 4.08GPTTSGGRR269 pKa = 11.84ASTPKK274 pKa = 10.8GGDD277 pKa = 3.07AAA279 pKa = 4.65
Molecular weight: 29.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2360 |
279 |
763 |
590.0 |
63.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.907 ± 0.926 | 1.186 ± 0.311 |
6.737 ± 0.19 | 4.449 ± 0.404 |
3.305 ± 0.459 | 7.119 ± 0.906 |
3.008 ± 0.204 | 3.771 ± 0.238 |
3.093 ± 0.631 | 9.153 ± 0.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.864 ± 0.343 | 2.542 ± 0.586 |
6.78 ± 0.496 | 2.5 ± 0.351 |
7.415 ± 0.297 | 7.373 ± 0.395 |
5.932 ± 0.383 | 8.22 ± 0.389 |
0.763 ± 0.063 | 2.881 ± 0.203 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
