
Nonomuraea fuscirosea
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiaceae; Nonomuraea
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
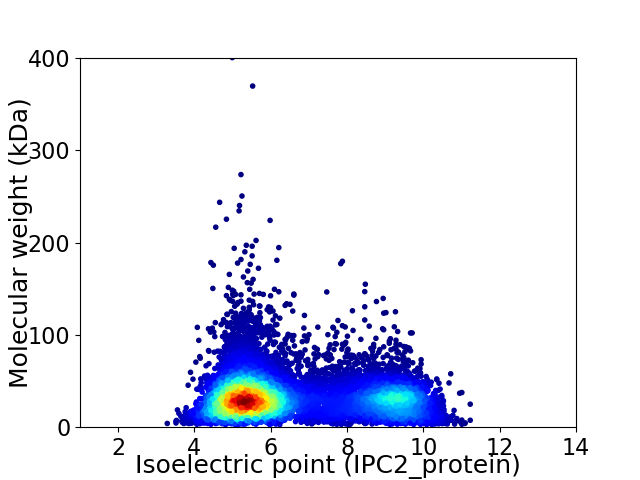
Virtual 2D-PAGE plot for 11276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0MPS6|A0A2T0MPS6_9ACTN Xylose isomerase OS=Nonomuraea fuscirosea OX=1291556 GN=xylA PE=3 SV=1
MM1 pKa = 7.1WWGTAIEE8 pKa = 4.68APDD11 pKa = 4.08PAALAQFYY19 pKa = 10.78SDD21 pKa = 3.91LLGWPIGHH29 pKa = 5.99QEE31 pKa = 4.08PGTTILAAPEE41 pKa = 3.74GSIYY45 pKa = 10.32IVFQQATDD53 pKa = 3.6YY54 pKa = 10.02QAPVWPPAAGQQRR67 pKa = 11.84PMMHH71 pKa = 7.14FDD73 pKa = 3.66FQVGDD78 pKa = 3.86LDD80 pKa = 4.06SAVAEE85 pKa = 4.3AVALGASVATEE96 pKa = 3.84QPQDD100 pKa = 3.36NVRR103 pKa = 11.84VLFDD107 pKa = 3.71PAGHH111 pKa = 6.94PFCLCRR117 pKa = 11.84DD118 pKa = 3.93DD119 pKa = 4.52GG120 pKa = 4.15
MM1 pKa = 7.1WWGTAIEE8 pKa = 4.68APDD11 pKa = 4.08PAALAQFYY19 pKa = 10.78SDD21 pKa = 3.91LLGWPIGHH29 pKa = 5.99QEE31 pKa = 4.08PGTTILAAPEE41 pKa = 3.74GSIYY45 pKa = 10.32IVFQQATDD53 pKa = 3.6YY54 pKa = 10.02QAPVWPPAAGQQRR67 pKa = 11.84PMMHH71 pKa = 7.14FDD73 pKa = 3.66FQVGDD78 pKa = 3.86LDD80 pKa = 4.06SAVAEE85 pKa = 4.3AVALGASVATEE96 pKa = 3.84QPQDD100 pKa = 3.36NVRR103 pKa = 11.84VLFDD107 pKa = 3.71PAGHH111 pKa = 6.94PFCLCRR117 pKa = 11.84DD118 pKa = 3.93DD119 pKa = 4.52GG120 pKa = 4.15
Molecular weight: 12.94 kDa
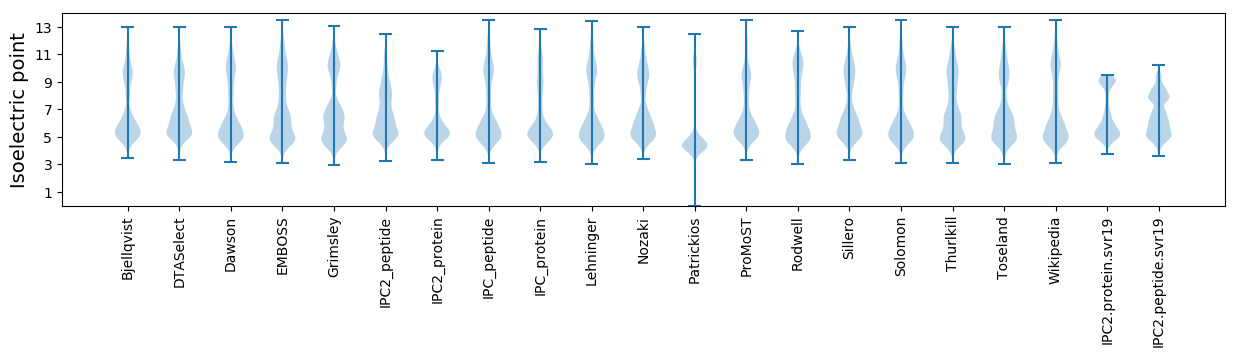
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0N9I1|A0A2T0N9I1_9ACTN Acetyl coenzyme A synthetase (ADP forming)-like protein OS=Nonomuraea fuscirosea OX=1291556 GN=B0I32_102427 PE=4 SV=1
MM1 pKa = 7.6ARR3 pKa = 11.84PPTAPVRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FPPPRR17 pKa = 11.84VRR19 pKa = 11.84ARR21 pKa = 11.84SPVRR25 pKa = 11.84VPPPQGAPPVAARR38 pKa = 11.84SSARR42 pKa = 11.84RR43 pKa = 11.84WRR45 pKa = 11.84CRR47 pKa = 11.84TSPSRR52 pKa = 11.84SLRR55 pKa = 11.84PPAARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GLRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84VSRR70 pKa = 11.84AARR73 pKa = 11.84PLVRR77 pKa = 11.84RR78 pKa = 11.84AASPLVPLRR87 pKa = 11.84ARR89 pKa = 11.84GPVPLLVRR97 pKa = 11.84APGPAPRR104 pKa = 11.84PVPGPRR110 pKa = 11.84AAAVASSRR118 pKa = 11.84LRR120 pKa = 11.84VPAAAVARR128 pKa = 11.84VRR130 pKa = 11.84DD131 pKa = 4.19RR132 pKa = 11.84VPSPARR138 pKa = 11.84VVRR141 pKa = 11.84VPATTRR147 pKa = 11.84SRR149 pKa = 11.84PTPAAWASPGRR160 pKa = 11.84RR161 pKa = 11.84VLAATARR168 pKa = 11.84AVAAPASVSVVTVRR182 pKa = 11.84RR183 pKa = 11.84ATARR187 pKa = 11.84CRR189 pKa = 11.84VRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VVAAARR199 pKa = 11.84AGPAAPVRR207 pKa = 11.84VLDD210 pKa = 4.1RR211 pKa = 11.84PPAARR216 pKa = 11.84VPVLVPVVPVLVARR230 pKa = 11.84GPTRR234 pKa = 3.29
MM1 pKa = 7.6ARR3 pKa = 11.84PPTAPVRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FPPPRR17 pKa = 11.84VRR19 pKa = 11.84ARR21 pKa = 11.84SPVRR25 pKa = 11.84VPPPQGAPPVAARR38 pKa = 11.84SSARR42 pKa = 11.84RR43 pKa = 11.84WRR45 pKa = 11.84CRR47 pKa = 11.84TSPSRR52 pKa = 11.84SLRR55 pKa = 11.84PPAARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GLRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84VSRR70 pKa = 11.84AARR73 pKa = 11.84PLVRR77 pKa = 11.84RR78 pKa = 11.84AASPLVPLRR87 pKa = 11.84ARR89 pKa = 11.84GPVPLLVRR97 pKa = 11.84APGPAPRR104 pKa = 11.84PVPGPRR110 pKa = 11.84AAAVASSRR118 pKa = 11.84LRR120 pKa = 11.84VPAAAVARR128 pKa = 11.84VRR130 pKa = 11.84DD131 pKa = 4.19RR132 pKa = 11.84VPSPARR138 pKa = 11.84VVRR141 pKa = 11.84VPATTRR147 pKa = 11.84SRR149 pKa = 11.84PTPAAWASPGRR160 pKa = 11.84RR161 pKa = 11.84VLAATARR168 pKa = 11.84AVAAPASVSVVTVRR182 pKa = 11.84RR183 pKa = 11.84ATARR187 pKa = 11.84CRR189 pKa = 11.84VRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VVAAARR199 pKa = 11.84AGPAAPVRR207 pKa = 11.84VLDD210 pKa = 4.1RR211 pKa = 11.84PPAARR216 pKa = 11.84VPVLVPVVPVLVARR230 pKa = 11.84GPTRR234 pKa = 3.29
Molecular weight: 24.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3590346 |
25 |
3748 |
318.4 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.349 ± 0.033 | 0.778 ± 0.007 |
5.733 ± 0.018 | 5.585 ± 0.021 |
2.814 ± 0.013 | 9.466 ± 0.023 |
2.223 ± 0.014 | 3.499 ± 0.015 |
1.987 ± 0.016 | 10.635 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.884 ± 0.008 | 1.753 ± 0.012 |
6.098 ± 0.019 | 2.704 ± 0.014 |
8.283 ± 0.026 | 5.023 ± 0.017 |
5.881 ± 0.021 | 8.578 ± 0.02 |
1.593 ± 0.01 | 2.134 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
