
Frankia sp. (strain EAN1pec)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Frankiales; Frankiaceae; Frankia; unclassified Frankia
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
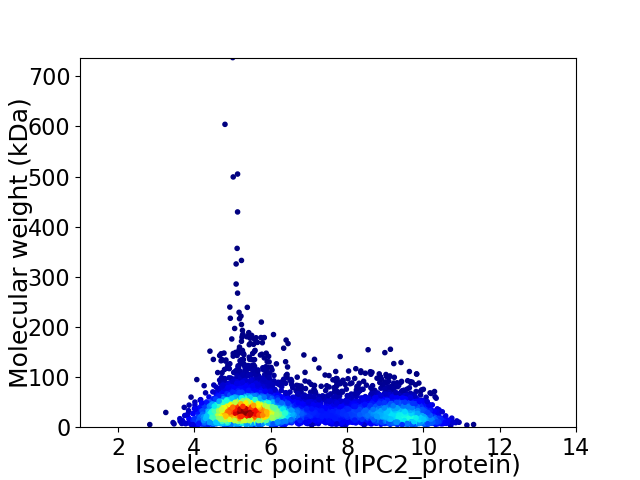
Virtual 2D-PAGE plot for 7133 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8LAM3|A8LAM3_FRASN 40-residue YVTN family beta-propeller repeat protein OS=Frankia sp. (strain EAN1pec) OX=298653 GN=Franean1_4419 PE=4 SV=1
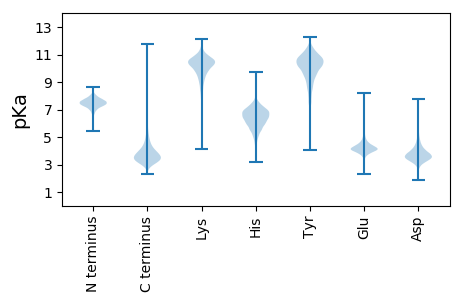
MM1 pKa = 7.87PEE3 pKa = 4.37DD4 pKa = 4.18SDD6 pKa = 5.32TILLDD11 pKa = 3.67YY12 pKa = 11.06ALSTDD17 pKa = 4.03PAPVPASTAQVVNQARR33 pKa = 11.84VNIGVSAEE41 pKa = 4.1TTVYY45 pKa = 10.43CSQIVIAVPIGTDD58 pKa = 3.46DD59 pKa = 3.7ASLYY63 pKa = 10.96ALAPAGSSNSNKK75 pKa = 9.21WAVTAQLRR83 pKa = 11.84KK84 pKa = 10.24GRR86 pKa = 11.84DD87 pKa = 2.96LWDD90 pKa = 4.41GLDD93 pKa = 3.5EE94 pKa = 4.8DD95 pKa = 3.96QDD97 pKa = 4.12YY98 pKa = 8.76STFVYY103 pKa = 10.59DD104 pKa = 4.36CVSSTDD110 pKa = 3.22YY111 pKa = 10.99RR112 pKa = 11.84VDD114 pKa = 3.4YY115 pKa = 10.96NLVFGVFGAVNQTVGDD131 pKa = 3.93AEE133 pKa = 4.12IAIQEE138 pKa = 4.28TSGTSSDD145 pKa = 3.39PTTFTLKK152 pKa = 10.1PAYY155 pKa = 10.19FPVRR159 pKa = 11.84KK160 pKa = 8.74EE161 pKa = 3.55WPAFYY166 pKa = 10.66LKK168 pKa = 10.83NLVAATLASPTVPATDD184 pKa = 4.11FSEE187 pKa = 4.53GDD189 pKa = 3.55PVRR192 pKa = 11.84LSWEE196 pKa = 4.1SNGTYY201 pKa = 9.92FQLFTKK207 pKa = 9.84GQSRR211 pKa = 11.84PVYY214 pKa = 10.11AGTATTYY221 pKa = 9.93TLEE224 pKa = 4.45EE225 pKa = 4.35GLSRR229 pKa = 11.84DD230 pKa = 3.32TAFVLTASMTGSPDD244 pKa = 3.06EE245 pKa = 5.18DD246 pKa = 4.05VPQGGYY252 pKa = 10.16EE253 pKa = 4.13PIYY256 pKa = 10.49LYY258 pKa = 10.83DD259 pKa = 4.25ALTVSVSNPVLTPSSVTVSGTLEE282 pKa = 3.97VSGTSYY288 pKa = 11.53LSTTEE293 pKa = 3.95TGSLSSSSVSVNGSLDD309 pKa = 3.46VDD311 pKa = 4.29GSTSLSDD318 pKa = 3.29TTVDD322 pKa = 4.05DD323 pKa = 4.19GLTVRR328 pKa = 11.84GSTEE332 pKa = 3.94LEE334 pKa = 3.85EE335 pKa = 4.34SLSVSGSTSLSDD347 pKa = 3.32ATVTGTLSGTGSASLSDD364 pKa = 3.32LTVRR368 pKa = 11.84GLYY371 pKa = 10.23GYY373 pKa = 9.6SGRR376 pKa = 11.84VEE378 pKa = 4.06LLGNANLVDD387 pKa = 3.57QGANLGQTGVYY398 pKa = 8.12TATDD402 pKa = 4.12GFAVAQVLTPSDD414 pKa = 3.67NSQASYY420 pKa = 10.84AYY422 pKa = 9.47GWIYY426 pKa = 10.69TSGTWFGVQGGTVGSFGPGWSDD448 pKa = 4.46FMNTNPNTITVPIPAGSYY466 pKa = 8.52WYY468 pKa = 10.35YY469 pKa = 11.2EE470 pKa = 4.09SFNPSNNQVYY480 pKa = 10.85SDD482 pKa = 3.65VQFWWFPLGSDD493 pKa = 3.54YY494 pKa = 11.29GATATYY500 pKa = 10.41RR501 pKa = 11.84KK502 pKa = 8.31LTAEE506 pKa = 4.07EE507 pKa = 4.14RR508 pKa = 11.84VAEE511 pKa = 4.15QVEE514 pKa = 4.66VPPARR519 pKa = 11.84PAVPDD524 pKa = 3.29IAAAVEE530 pKa = 4.05RR531 pKa = 11.84RR532 pKa = 11.84EE533 pKa = 3.77NAAAAFVEE541 pKa = 4.42RR542 pKa = 11.84LAAALEE548 pKa = 4.14ADD550 pKa = 3.62LAEE553 pKa = 4.52EE554 pKa = 4.28VRR556 pKa = 11.84ADD558 pKa = 3.59LTRR561 pKa = 11.84LLNQHH566 pKa = 6.3
MM1 pKa = 7.87PEE3 pKa = 4.37DD4 pKa = 4.18SDD6 pKa = 5.32TILLDD11 pKa = 3.67YY12 pKa = 11.06ALSTDD17 pKa = 4.03PAPVPASTAQVVNQARR33 pKa = 11.84VNIGVSAEE41 pKa = 4.1TTVYY45 pKa = 10.43CSQIVIAVPIGTDD58 pKa = 3.46DD59 pKa = 3.7ASLYY63 pKa = 10.96ALAPAGSSNSNKK75 pKa = 9.21WAVTAQLRR83 pKa = 11.84KK84 pKa = 10.24GRR86 pKa = 11.84DD87 pKa = 2.96LWDD90 pKa = 4.41GLDD93 pKa = 3.5EE94 pKa = 4.8DD95 pKa = 3.96QDD97 pKa = 4.12YY98 pKa = 8.76STFVYY103 pKa = 10.59DD104 pKa = 4.36CVSSTDD110 pKa = 3.22YY111 pKa = 10.99RR112 pKa = 11.84VDD114 pKa = 3.4YY115 pKa = 10.96NLVFGVFGAVNQTVGDD131 pKa = 3.93AEE133 pKa = 4.12IAIQEE138 pKa = 4.28TSGTSSDD145 pKa = 3.39PTTFTLKK152 pKa = 10.1PAYY155 pKa = 10.19FPVRR159 pKa = 11.84KK160 pKa = 8.74EE161 pKa = 3.55WPAFYY166 pKa = 10.66LKK168 pKa = 10.83NLVAATLASPTVPATDD184 pKa = 4.11FSEE187 pKa = 4.53GDD189 pKa = 3.55PVRR192 pKa = 11.84LSWEE196 pKa = 4.1SNGTYY201 pKa = 9.92FQLFTKK207 pKa = 9.84GQSRR211 pKa = 11.84PVYY214 pKa = 10.11AGTATTYY221 pKa = 9.93TLEE224 pKa = 4.45EE225 pKa = 4.35GLSRR229 pKa = 11.84DD230 pKa = 3.32TAFVLTASMTGSPDD244 pKa = 3.06EE245 pKa = 5.18DD246 pKa = 4.05VPQGGYY252 pKa = 10.16EE253 pKa = 4.13PIYY256 pKa = 10.49LYY258 pKa = 10.83DD259 pKa = 4.25ALTVSVSNPVLTPSSVTVSGTLEE282 pKa = 3.97VSGTSYY288 pKa = 11.53LSTTEE293 pKa = 3.95TGSLSSSSVSVNGSLDD309 pKa = 3.46VDD311 pKa = 4.29GSTSLSDD318 pKa = 3.29TTVDD322 pKa = 4.05DD323 pKa = 4.19GLTVRR328 pKa = 11.84GSTEE332 pKa = 3.94LEE334 pKa = 3.85EE335 pKa = 4.34SLSVSGSTSLSDD347 pKa = 3.32ATVTGTLSGTGSASLSDD364 pKa = 3.32LTVRR368 pKa = 11.84GLYY371 pKa = 10.23GYY373 pKa = 9.6SGRR376 pKa = 11.84VEE378 pKa = 4.06LLGNANLVDD387 pKa = 3.57QGANLGQTGVYY398 pKa = 8.12TATDD402 pKa = 4.12GFAVAQVLTPSDD414 pKa = 3.67NSQASYY420 pKa = 10.84AYY422 pKa = 9.47GWIYY426 pKa = 10.69TSGTWFGVQGGTVGSFGPGWSDD448 pKa = 4.46FMNTNPNTITVPIPAGSYY466 pKa = 8.52WYY468 pKa = 10.35YY469 pKa = 11.2EE470 pKa = 4.09SFNPSNNQVYY480 pKa = 10.85SDD482 pKa = 3.65VQFWWFPLGSDD493 pKa = 3.54YY494 pKa = 11.29GATATYY500 pKa = 10.41RR501 pKa = 11.84KK502 pKa = 8.31LTAEE506 pKa = 4.07EE507 pKa = 4.14RR508 pKa = 11.84VAEE511 pKa = 4.15QVEE514 pKa = 4.66VPPARR519 pKa = 11.84PAVPDD524 pKa = 3.29IAAAVEE530 pKa = 4.05RR531 pKa = 11.84RR532 pKa = 11.84EE533 pKa = 3.77NAAAAFVEE541 pKa = 4.42RR542 pKa = 11.84LAAALEE548 pKa = 4.14ADD550 pKa = 3.62LAEE553 pKa = 4.52EE554 pKa = 4.28VRR556 pKa = 11.84ADD558 pKa = 3.59LTRR561 pKa = 11.84LLNQHH566 pKa = 6.3
Molecular weight: 60.01 kDa
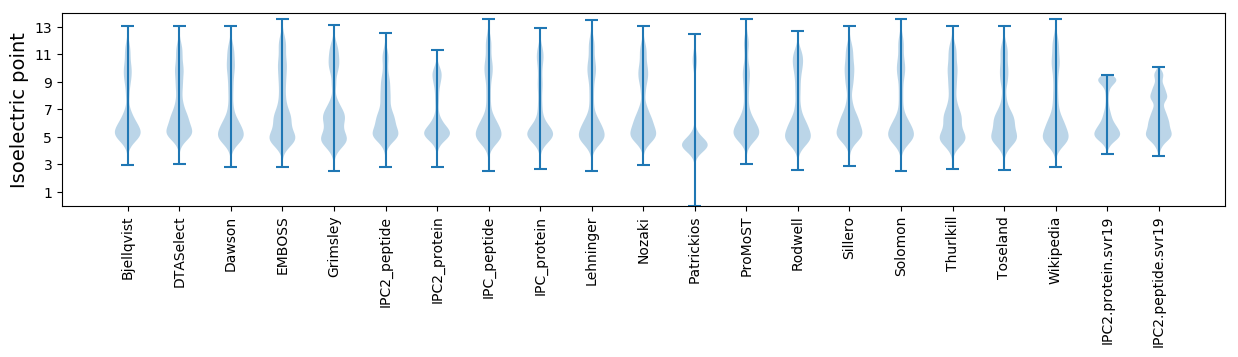
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8L989|A8L989_FRASN Uncharacterized protein OS=Frankia sp. (strain EAN1pec) OX=298653 GN=Franean1_2805 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 5.69GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.93GRR40 pKa = 11.84SQLSAA45 pKa = 3.6
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 5.69GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.93GRR40 pKa = 11.84SQLSAA45 pKa = 3.6
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2476290 |
26 |
6999 |
347.2 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.01 ± 0.053 | 0.797 ± 0.008 |
6.129 ± 0.02 | 5.064 ± 0.026 |
2.631 ± 0.016 | 9.698 ± 0.038 |
2.226 ± 0.014 | 3.358 ± 0.019 |
1.396 ± 0.015 | 10.048 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.633 ± 0.013 | 1.67 ± 0.013 |
6.705 ± 0.032 | 2.585 ± 0.016 |
8.802 ± 0.03 | 5.283 ± 0.022 |
6.098 ± 0.021 | 8.584 ± 0.03 |
1.451 ± 0.012 | 1.832 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
