
Mycobacterium sp. ACS4331
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; unclassified Mycobacterium
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
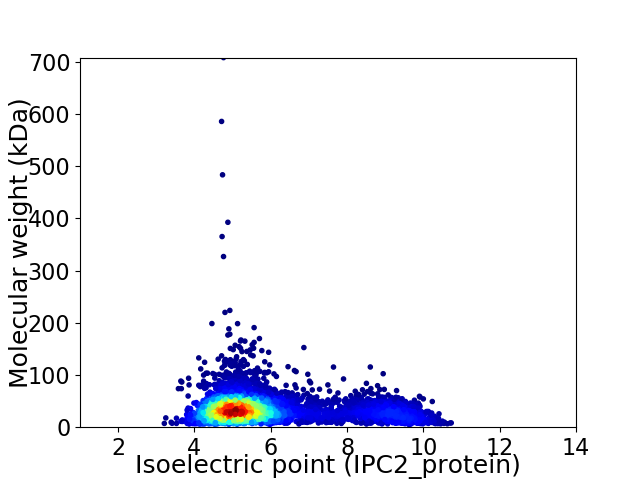
Virtual 2D-PAGE plot for 5193 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A1Y0W3|A0A1A1Y0W3_9MYCO 50S ribosomal protein L21 OS=Mycobacterium sp. ACS4331 OX=1834121 GN=rplU PE=3 SV=1
MM1 pKa = 7.11PTEE4 pKa = 4.52APLDD8 pKa = 3.88LVLLAWTRR16 pKa = 11.84RR17 pKa = 11.84QNALTAAGVTRR28 pKa = 11.84ATDD31 pKa = 2.93THH33 pKa = 6.02QRR35 pKa = 11.84GQIAEE40 pKa = 4.28QINSIAMLSTAQAAAQLANAYY61 pKa = 9.44VAEE64 pKa = 5.21DD65 pKa = 4.09DD66 pKa = 4.77PEE68 pKa = 5.57LGNIGSGNVGGGNQGSGNTGSGNTGNNNGGDD99 pKa = 4.0GNIGDD104 pKa = 4.3GNAGNGNQGTGNAGDD119 pKa = 4.16GNTGNNNDD127 pKa = 3.6GNGNTGSGNIGDD139 pKa = 4.33GNQGGNNQGHH149 pKa = 6.75GNTGNANVGDD159 pKa = 4.18GNSGNGNAGDD169 pKa = 4.07GNTGVGNAGDD179 pKa = 4.3GNTGTGNAGDD189 pKa = 4.43GNTGDD194 pKa = 4.43SNPGSGNQGAGNVGDD209 pKa = 4.33GNQGGDD215 pKa = 3.54NQGHH219 pKa = 6.59GNTGNANVGDD229 pKa = 4.32GNSGTGNVGDD239 pKa = 4.31GNTGNGNAGDD249 pKa = 4.18GNQGDD254 pKa = 4.77DD255 pKa = 3.7NVGDD259 pKa = 4.22GNSGTGNTGHH269 pKa = 6.86GNQGSTNAGDD279 pKa = 4.71GNIGDD284 pKa = 4.36NNAGSGNAGDD294 pKa = 5.11LNTGSGNAGSGNSGSGNSGSGNTGDD319 pKa = 4.99GNQGHH324 pKa = 6.49NNAGDD329 pKa = 3.91GNTGTGNAGDD339 pKa = 4.36GNIGDD344 pKa = 4.86HH345 pKa = 5.77NTGSGNDD352 pKa = 3.54GDD354 pKa = 5.18GNTGHH359 pKa = 7.03GNTGNTNTGHH369 pKa = 6.27GNSGSGNQGDD379 pKa = 5.03GNIGDD384 pKa = 4.0NNLPGAHH391 pKa = 6.75GNTGDD396 pKa = 4.75DD397 pKa = 3.58NTGSGNRR404 pKa = 11.84GDD406 pKa = 5.43DD407 pKa = 3.46NTGSGNIGDD416 pKa = 4.36GNGGDD421 pKa = 4.57GNIGDD426 pKa = 4.92DD427 pKa = 3.74NSGSGNIGDD436 pKa = 4.58HH437 pKa = 6.45NSGSGNQGSGNSGGGNRR454 pKa = 11.84GDD456 pKa = 4.37DD457 pKa = 3.53NSGSGNIGDD466 pKa = 4.7HH467 pKa = 5.47NTGHH471 pKa = 6.88GNQGDD476 pKa = 4.08NNSGSGNSGTGLGGGDD492 pKa = 3.61NNSGSGNQGSGNSGSGNIGNDD513 pKa = 3.22NTGHH517 pKa = 6.77ANQGGDD523 pKa = 3.23NSGHH527 pKa = 6.72ANIGDD532 pKa = 3.81NNQGSGNIGSGNSGSGNIGSGNQGSGNIGADD563 pKa = 3.32NQGGGNTGNANPGWGNIGNNNIGLGNKK590 pKa = 9.17GNNNVGFGNTGNNNFGIGLVGDD612 pKa = 3.78NLIGIGGFNSGWFNLGLGNAGNFNIGFGNSGVGNIGFGNSGNYY655 pKa = 10.25NIGFFNSGDD664 pKa = 3.71GNVGFWNSGSGNTGFWNSGKK684 pKa = 10.71ANTGFYY690 pKa = 10.97NSGNTNTGFYY700 pKa = 10.93NSGNTNTGFWNSGDD714 pKa = 3.73TNTGFWNSGNTNTGCYY730 pKa = 10.18NSGDD734 pKa = 4.05TNTGCWNSGNTNTGFSNSGNTNTGKK759 pKa = 10.15HH760 pKa = 5.29NSGGTNTGNYY770 pKa = 8.69NAGGTNTGHH779 pKa = 6.76HH780 pKa = 6.13NAGGTNTGSYY790 pKa = 10.0NAGGTNTGNRR800 pKa = 11.84NAGGTNYY807 pKa = 10.61GDD809 pKa = 3.88YY810 pKa = 11.23NSGGTNTGDD819 pKa = 3.66YY820 pKa = 11.11NSGGTNTGSYY830 pKa = 10.89NSGGTNTGSYY840 pKa = 10.89NSGGTNTGDD849 pKa = 3.66YY850 pKa = 11.11NSGGTNTGHH859 pKa = 6.08YY860 pKa = 10.35NSGGTNTGDD869 pKa = 3.66YY870 pKa = 11.11NSGGTNTGHH879 pKa = 7.34DD880 pKa = 3.7NSGSTNTGNFNAGNTNTGNLNSGNTNTGHH909 pKa = 7.05LNSGDD914 pKa = 3.91TNTGDD919 pKa = 4.0LNSGNTNTGNANSGNTNRR937 pKa = 11.84CDD939 pKa = 3.54HH940 pKa = 6.58NSGNTNTGDD949 pKa = 3.66RR950 pKa = 11.84NSGDD954 pKa = 3.9TNTGNYY960 pKa = 10.08NSGSGNIGDD969 pKa = 4.58HH970 pKa = 6.67NSGSGNNGDD979 pKa = 3.87TNTGHH984 pKa = 7.37RR985 pKa = 11.84KK986 pKa = 7.79TGG988 pKa = 3.24
MM1 pKa = 7.11PTEE4 pKa = 4.52APLDD8 pKa = 3.88LVLLAWTRR16 pKa = 11.84RR17 pKa = 11.84QNALTAAGVTRR28 pKa = 11.84ATDD31 pKa = 2.93THH33 pKa = 6.02QRR35 pKa = 11.84GQIAEE40 pKa = 4.28QINSIAMLSTAQAAAQLANAYY61 pKa = 9.44VAEE64 pKa = 5.21DD65 pKa = 4.09DD66 pKa = 4.77PEE68 pKa = 5.57LGNIGSGNVGGGNQGSGNTGSGNTGNNNGGDD99 pKa = 4.0GNIGDD104 pKa = 4.3GNAGNGNQGTGNAGDD119 pKa = 4.16GNTGNNNDD127 pKa = 3.6GNGNTGSGNIGDD139 pKa = 4.33GNQGGNNQGHH149 pKa = 6.75GNTGNANVGDD159 pKa = 4.18GNSGNGNAGDD169 pKa = 4.07GNTGVGNAGDD179 pKa = 4.3GNTGTGNAGDD189 pKa = 4.43GNTGDD194 pKa = 4.43SNPGSGNQGAGNVGDD209 pKa = 4.33GNQGGDD215 pKa = 3.54NQGHH219 pKa = 6.59GNTGNANVGDD229 pKa = 4.32GNSGTGNVGDD239 pKa = 4.31GNTGNGNAGDD249 pKa = 4.18GNQGDD254 pKa = 4.77DD255 pKa = 3.7NVGDD259 pKa = 4.22GNSGTGNTGHH269 pKa = 6.86GNQGSTNAGDD279 pKa = 4.71GNIGDD284 pKa = 4.36NNAGSGNAGDD294 pKa = 5.11LNTGSGNAGSGNSGSGNSGSGNTGDD319 pKa = 4.99GNQGHH324 pKa = 6.49NNAGDD329 pKa = 3.91GNTGTGNAGDD339 pKa = 4.36GNIGDD344 pKa = 4.86HH345 pKa = 5.77NTGSGNDD352 pKa = 3.54GDD354 pKa = 5.18GNTGHH359 pKa = 7.03GNTGNTNTGHH369 pKa = 6.27GNSGSGNQGDD379 pKa = 5.03GNIGDD384 pKa = 4.0NNLPGAHH391 pKa = 6.75GNTGDD396 pKa = 4.75DD397 pKa = 3.58NTGSGNRR404 pKa = 11.84GDD406 pKa = 5.43DD407 pKa = 3.46NTGSGNIGDD416 pKa = 4.36GNGGDD421 pKa = 4.57GNIGDD426 pKa = 4.92DD427 pKa = 3.74NSGSGNIGDD436 pKa = 4.58HH437 pKa = 6.45NSGSGNQGSGNSGGGNRR454 pKa = 11.84GDD456 pKa = 4.37DD457 pKa = 3.53NSGSGNIGDD466 pKa = 4.7HH467 pKa = 5.47NTGHH471 pKa = 6.88GNQGDD476 pKa = 4.08NNSGSGNSGTGLGGGDD492 pKa = 3.61NNSGSGNQGSGNSGSGNIGNDD513 pKa = 3.22NTGHH517 pKa = 6.77ANQGGDD523 pKa = 3.23NSGHH527 pKa = 6.72ANIGDD532 pKa = 3.81NNQGSGNIGSGNSGSGNIGSGNQGSGNIGADD563 pKa = 3.32NQGGGNTGNANPGWGNIGNNNIGLGNKK590 pKa = 9.17GNNNVGFGNTGNNNFGIGLVGDD612 pKa = 3.78NLIGIGGFNSGWFNLGLGNAGNFNIGFGNSGVGNIGFGNSGNYY655 pKa = 10.25NIGFFNSGDD664 pKa = 3.71GNVGFWNSGSGNTGFWNSGKK684 pKa = 10.71ANTGFYY690 pKa = 10.97NSGNTNTGFYY700 pKa = 10.93NSGNTNTGFWNSGDD714 pKa = 3.73TNTGFWNSGNTNTGCYY730 pKa = 10.18NSGDD734 pKa = 4.05TNTGCWNSGNTNTGFSNSGNTNTGKK759 pKa = 10.15HH760 pKa = 5.29NSGGTNTGNYY770 pKa = 8.69NAGGTNTGHH779 pKa = 6.76HH780 pKa = 6.13NAGGTNTGSYY790 pKa = 10.0NAGGTNTGNRR800 pKa = 11.84NAGGTNYY807 pKa = 10.61GDD809 pKa = 3.88YY810 pKa = 11.23NSGGTNTGDD819 pKa = 3.66YY820 pKa = 11.11NSGGTNTGSYY830 pKa = 10.89NSGGTNTGSYY840 pKa = 10.89NSGGTNTGDD849 pKa = 3.66YY850 pKa = 11.11NSGGTNTGHH859 pKa = 6.08YY860 pKa = 10.35NSGGTNTGDD869 pKa = 3.66YY870 pKa = 11.11NSGGTNTGHH879 pKa = 7.34DD880 pKa = 3.7NSGSTNTGNFNAGNTNTGNLNSGNTNTGHH909 pKa = 7.05LNSGDD914 pKa = 3.91TNTGDD919 pKa = 4.0LNSGNTNTGNANSGNTNRR937 pKa = 11.84CDD939 pKa = 3.54HH940 pKa = 6.58NSGNTNTGDD949 pKa = 3.66RR950 pKa = 11.84NSGDD954 pKa = 3.9TNTGNYY960 pKa = 10.08NSGSGNIGDD969 pKa = 4.58HH970 pKa = 6.67NSGSGNNGDD979 pKa = 3.87TNTGHH984 pKa = 7.37RR985 pKa = 11.84KK986 pKa = 7.79TGG988 pKa = 3.24
Molecular weight: 93.78 kDa
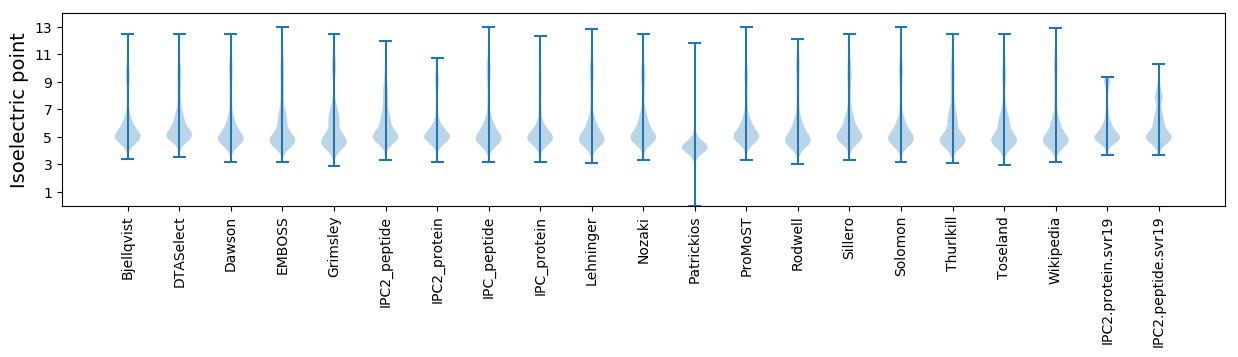
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A1WU81|A0A1A1WU81_9MYCO ATP synthase subunit alpha OS=Mycobacterium sp. ACS4331 OX=1834121 GN=atpA PE=3 SV=1
MM1 pKa = 7.3AQHH4 pKa = 6.38KK5 pKa = 10.27RR6 pKa = 11.84NLAKK10 pKa = 10.31RR11 pKa = 11.84VGHH14 pKa = 6.6RR15 pKa = 11.84SSAGPVGLGLGTPLHH30 pKa = 6.13SVEE33 pKa = 4.83NVATSPPVAHH43 pKa = 7.6DD44 pKa = 3.63STSVWGRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VLLLNSTYY61 pKa = 10.82EE62 pKa = 4.11PLTALPMRR70 pKa = 11.84RR71 pKa = 11.84AIVMLLCGKK80 pKa = 10.53ADD82 pKa = 3.62VVHH85 pKa = 7.72DD86 pKa = 4.63DD87 pKa = 3.59PNGPVIHH94 pKa = 6.52SATRR98 pKa = 11.84AIAVPSVIRR107 pKa = 11.84LRR109 pKa = 11.84SYY111 pKa = 10.66VRR113 pKa = 11.84VPYY116 pKa = 10.05RR117 pKa = 11.84ARR119 pKa = 11.84VPMTRR124 pKa = 11.84AALMHH129 pKa = 6.99RR130 pKa = 11.84DD131 pKa = 3.56RR132 pKa = 11.84FRR134 pKa = 11.84CAYY137 pKa = 9.43CGSKK141 pKa = 10.56ADD143 pKa = 4.08TVDD146 pKa = 3.26HH147 pKa = 5.51VVPRR151 pKa = 11.84SRR153 pKa = 11.84GGEE156 pKa = 4.13HH157 pKa = 4.92TWEE160 pKa = 4.1NCVACCSTCNHH171 pKa = 6.33RR172 pKa = 11.84KK173 pKa = 9.55ADD175 pKa = 4.13RR176 pKa = 11.84LLSEE180 pKa = 5.25LGWSLRR186 pKa = 11.84RR187 pKa = 11.84APMPPKK193 pKa = 8.9GQHH196 pKa = 4.68WRR198 pKa = 11.84LLSTVKK204 pKa = 10.41EE205 pKa = 4.26LDD207 pKa = 4.0PAWVRR212 pKa = 11.84YY213 pKa = 9.55LGEE216 pKa = 4.14GAAA219 pKa = 4.63
MM1 pKa = 7.3AQHH4 pKa = 6.38KK5 pKa = 10.27RR6 pKa = 11.84NLAKK10 pKa = 10.31RR11 pKa = 11.84VGHH14 pKa = 6.6RR15 pKa = 11.84SSAGPVGLGLGTPLHH30 pKa = 6.13SVEE33 pKa = 4.83NVATSPPVAHH43 pKa = 7.6DD44 pKa = 3.63STSVWGRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VLLLNSTYY61 pKa = 10.82EE62 pKa = 4.11PLTALPMRR70 pKa = 11.84RR71 pKa = 11.84AIVMLLCGKK80 pKa = 10.53ADD82 pKa = 3.62VVHH85 pKa = 7.72DD86 pKa = 4.63DD87 pKa = 3.59PNGPVIHH94 pKa = 6.52SATRR98 pKa = 11.84AIAVPSVIRR107 pKa = 11.84LRR109 pKa = 11.84SYY111 pKa = 10.66VRR113 pKa = 11.84VPYY116 pKa = 10.05RR117 pKa = 11.84ARR119 pKa = 11.84VPMTRR124 pKa = 11.84AALMHH129 pKa = 6.99RR130 pKa = 11.84DD131 pKa = 3.56RR132 pKa = 11.84FRR134 pKa = 11.84CAYY137 pKa = 9.43CGSKK141 pKa = 10.56ADD143 pKa = 4.08TVDD146 pKa = 3.26HH147 pKa = 5.51VVPRR151 pKa = 11.84SRR153 pKa = 11.84GGEE156 pKa = 4.13HH157 pKa = 4.92TWEE160 pKa = 4.1NCVACCSTCNHH171 pKa = 6.33RR172 pKa = 11.84KK173 pKa = 9.55ADD175 pKa = 4.13RR176 pKa = 11.84LLSEE180 pKa = 5.25LGWSLRR186 pKa = 11.84RR187 pKa = 11.84APMPPKK193 pKa = 8.9GQHH196 pKa = 4.68WRR198 pKa = 11.84LLSTVKK204 pKa = 10.41EE205 pKa = 4.26LDD207 pKa = 4.0PAWVRR212 pKa = 11.84YY213 pKa = 9.55LGEE216 pKa = 4.14GAAA219 pKa = 4.63
Molecular weight: 24.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1739715 |
37 |
6600 |
335.0 |
35.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.06 ± 0.047 | 0.8 ± 0.008 |
6.405 ± 0.025 | 5.364 ± 0.031 |
3.075 ± 0.019 | 8.942 ± 0.051 |
2.202 ± 0.014 | 4.211 ± 0.022 |
2.036 ± 0.024 | 9.838 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.013 | 2.207 ± 0.023 |
5.815 ± 0.031 | 2.941 ± 0.015 |
7.177 ± 0.034 | 5.369 ± 0.019 |
6.102 ± 0.024 | 8.878 ± 0.034 |
1.482 ± 0.016 | 2.066 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
