
Phytophthora cactorum
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
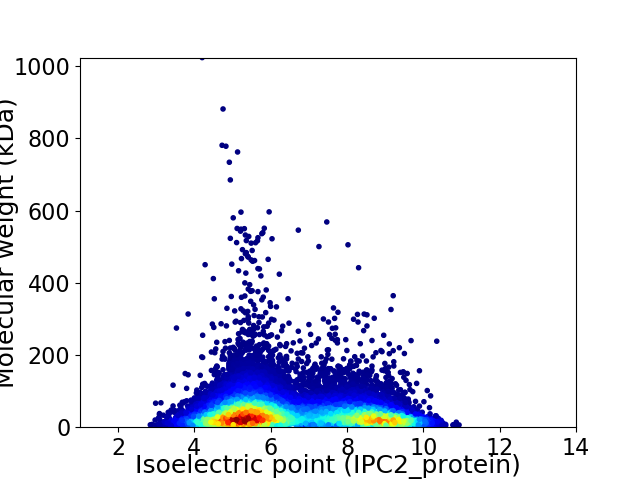
Virtual 2D-PAGE plot for 23398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A329RW33|A0A329RW33_9STRA Uncharacterized protein OS=Phytophthora cactorum OX=29920 GN=PC110_g14849 PE=4 SV=1
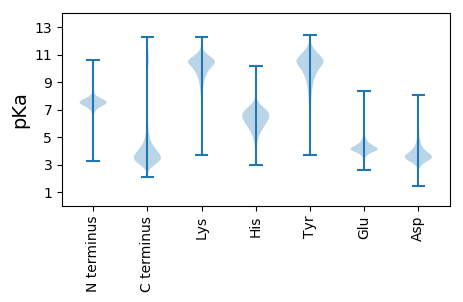
MM1 pKa = 7.77KK2 pKa = 10.45SPSSLVLLATAATFAFADD20 pKa = 3.49AGGIAAEE27 pKa = 4.45YY28 pKa = 8.03FTGDD32 pKa = 2.89GTAYY36 pKa = 9.79TLGDD40 pKa = 3.74TSSGNCNMMSALNFATTDD58 pKa = 3.58YY59 pKa = 11.49AALNNDD65 pKa = 3.04QWSGLQNCGRR75 pKa = 11.84CAEE78 pKa = 4.27VSCADD83 pKa = 4.19SRR85 pKa = 11.84CSDD88 pKa = 3.22QTKK91 pKa = 10.46SVVVQILDD99 pKa = 3.38RR100 pKa = 11.84CPEE103 pKa = 4.23CKK105 pKa = 10.06QGDD108 pKa = 4.41LDD110 pKa = 4.8LSPSVFKK117 pKa = 10.34TLTGSHH123 pKa = 7.3PSRR126 pKa = 11.84YY127 pKa = 7.04TIKK130 pKa = 10.03WKK132 pKa = 10.51FVDD135 pKa = 4.75CPVAGNVNYY144 pKa = 10.11CLKK147 pKa = 10.76GGSNNYY153 pKa = 5.91WTAIQPTNVATGVKK167 pKa = 9.46SLKK170 pKa = 10.43INGQATKK177 pKa = 9.46MLDD180 pKa = 2.98GAYY183 pKa = 10.49YY184 pKa = 10.59YY185 pKa = 11.17LLDD188 pKa = 4.53GAGTSQTDD196 pKa = 3.67LSSVSVSLTDD206 pKa = 3.33INGVSITDD214 pKa = 3.84KK215 pKa = 10.81VSLSAGSCAEE225 pKa = 4.29GSHH228 pKa = 5.46QFPSGGSPSKK238 pKa = 10.62PSTNTSTPTGAPKK251 pKa = 9.11PTTATPTMAPTKK263 pKa = 10.48VPTVTTKK270 pKa = 10.14ATTPTTPTPSQEE282 pKa = 4.25QQTPAPVPTTSTPMTDD298 pKa = 3.41TPASTTPTSTTQSGSGDD315 pKa = 3.85DD316 pKa = 3.98ASQTAQQDD324 pKa = 3.46LGSASRR330 pKa = 11.84NDD332 pKa = 4.08DD333 pKa = 3.41ATQSSPQASSSGNGDD348 pKa = 3.32SPEE351 pKa = 3.85QSIEE355 pKa = 3.79QDD357 pKa = 3.23PGSEE361 pKa = 4.23SNDD364 pKa = 3.18VEE366 pKa = 4.35TQAPITQQSSSSSEE380 pKa = 3.74GDD382 pKa = 3.06EE383 pKa = 4.33TQQQSSTDD391 pKa = 3.7GEE393 pKa = 4.72TSTEE397 pKa = 4.21SPDD400 pKa = 3.45VPATVAPSTPFSSASQQSGSGVDD423 pKa = 3.79SEE425 pKa = 5.17TEE427 pKa = 3.9ASSSASTYY435 pKa = 8.96MDD437 pKa = 4.46CGSSSHH443 pKa = 6.99PSEE446 pKa = 4.09QTPSSSNNDD455 pKa = 3.21NPSQEE460 pKa = 4.36SQQSAEE466 pKa = 3.82QDD468 pKa = 3.68LNSASVDD475 pKa = 3.43GNTDD479 pKa = 2.61NSTPIQQSGSTSDD492 pKa = 3.48NDD494 pKa = 3.7GTQVTQQDD502 pKa = 4.07PDD504 pKa = 3.92SVDD507 pKa = 4.21SEE509 pKa = 4.68DD510 pKa = 3.63STVDD514 pKa = 3.34NASSAVGADD523 pKa = 2.84VDD525 pKa = 4.22MVISQSGSADD535 pKa = 3.46QNDD538 pKa = 4.34EE539 pKa = 4.28NPVQQTSSDD548 pKa = 3.5VGADD552 pKa = 3.51TDD554 pKa = 4.63SEE556 pKa = 4.72TPEE559 pKa = 3.82QTSNEE564 pKa = 4.21DD565 pKa = 3.81GEE567 pKa = 4.93TEE569 pKa = 4.26TPQQDD574 pKa = 3.22SSSANTEE581 pKa = 3.85TTEE584 pKa = 4.24IPSSTGTANCPMM596 pKa = 5.11
MM1 pKa = 7.77KK2 pKa = 10.45SPSSLVLLATAATFAFADD20 pKa = 3.49AGGIAAEE27 pKa = 4.45YY28 pKa = 8.03FTGDD32 pKa = 2.89GTAYY36 pKa = 9.79TLGDD40 pKa = 3.74TSSGNCNMMSALNFATTDD58 pKa = 3.58YY59 pKa = 11.49AALNNDD65 pKa = 3.04QWSGLQNCGRR75 pKa = 11.84CAEE78 pKa = 4.27VSCADD83 pKa = 4.19SRR85 pKa = 11.84CSDD88 pKa = 3.22QTKK91 pKa = 10.46SVVVQILDD99 pKa = 3.38RR100 pKa = 11.84CPEE103 pKa = 4.23CKK105 pKa = 10.06QGDD108 pKa = 4.41LDD110 pKa = 4.8LSPSVFKK117 pKa = 10.34TLTGSHH123 pKa = 7.3PSRR126 pKa = 11.84YY127 pKa = 7.04TIKK130 pKa = 10.03WKK132 pKa = 10.51FVDD135 pKa = 4.75CPVAGNVNYY144 pKa = 10.11CLKK147 pKa = 10.76GGSNNYY153 pKa = 5.91WTAIQPTNVATGVKK167 pKa = 9.46SLKK170 pKa = 10.43INGQATKK177 pKa = 9.46MLDD180 pKa = 2.98GAYY183 pKa = 10.49YY184 pKa = 10.59YY185 pKa = 11.17LLDD188 pKa = 4.53GAGTSQTDD196 pKa = 3.67LSSVSVSLTDD206 pKa = 3.33INGVSITDD214 pKa = 3.84KK215 pKa = 10.81VSLSAGSCAEE225 pKa = 4.29GSHH228 pKa = 5.46QFPSGGSPSKK238 pKa = 10.62PSTNTSTPTGAPKK251 pKa = 9.11PTTATPTMAPTKK263 pKa = 10.48VPTVTTKK270 pKa = 10.14ATTPTTPTPSQEE282 pKa = 4.25QQTPAPVPTTSTPMTDD298 pKa = 3.41TPASTTPTSTTQSGSGDD315 pKa = 3.85DD316 pKa = 3.98ASQTAQQDD324 pKa = 3.46LGSASRR330 pKa = 11.84NDD332 pKa = 4.08DD333 pKa = 3.41ATQSSPQASSSGNGDD348 pKa = 3.32SPEE351 pKa = 3.85QSIEE355 pKa = 3.79QDD357 pKa = 3.23PGSEE361 pKa = 4.23SNDD364 pKa = 3.18VEE366 pKa = 4.35TQAPITQQSSSSSEE380 pKa = 3.74GDD382 pKa = 3.06EE383 pKa = 4.33TQQQSSTDD391 pKa = 3.7GEE393 pKa = 4.72TSTEE397 pKa = 4.21SPDD400 pKa = 3.45VPATVAPSTPFSSASQQSGSGVDD423 pKa = 3.79SEE425 pKa = 5.17TEE427 pKa = 3.9ASSSASTYY435 pKa = 8.96MDD437 pKa = 4.46CGSSSHH443 pKa = 6.99PSEE446 pKa = 4.09QTPSSSNNDD455 pKa = 3.21NPSQEE460 pKa = 4.36SQQSAEE466 pKa = 3.82QDD468 pKa = 3.68LNSASVDD475 pKa = 3.43GNTDD479 pKa = 2.61NSTPIQQSGSTSDD492 pKa = 3.48NDD494 pKa = 3.7GTQVTQQDD502 pKa = 4.07PDD504 pKa = 3.92SVDD507 pKa = 4.21SEE509 pKa = 4.68DD510 pKa = 3.63STVDD514 pKa = 3.34NASSAVGADD523 pKa = 2.84VDD525 pKa = 4.22MVISQSGSADD535 pKa = 3.46QNDD538 pKa = 4.34EE539 pKa = 4.28NPVQQTSSDD548 pKa = 3.5VGADD552 pKa = 3.51TDD554 pKa = 4.63SEE556 pKa = 4.72TPEE559 pKa = 3.82QTSNEE564 pKa = 4.21DD565 pKa = 3.81GEE567 pKa = 4.93TEE569 pKa = 4.26TPQQDD574 pKa = 3.22SSSANTEE581 pKa = 3.85TTEE584 pKa = 4.24IPSSTGTANCPMM596 pKa = 5.11
Molecular weight: 61.03 kDa
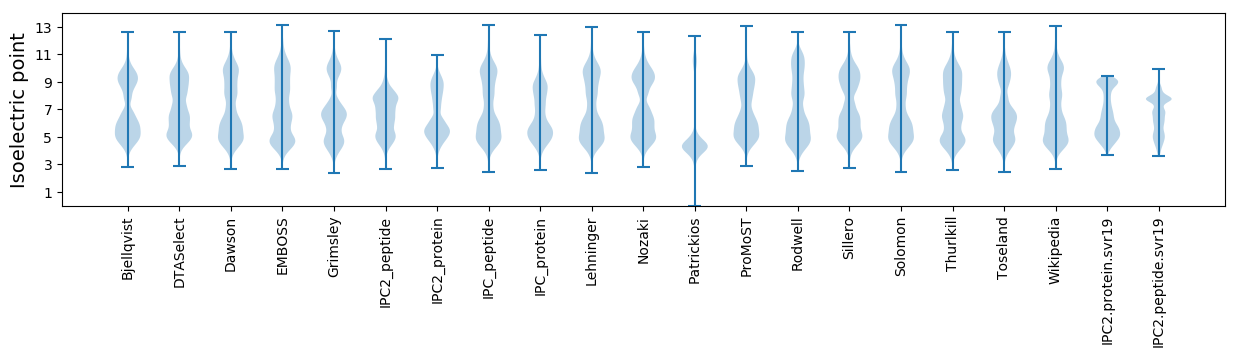
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A329S3I7|A0A329S3I7_9STRA Dymeclin OS=Phytophthora cactorum OX=29920 GN=PC110_g12338 PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 10.19RR3 pKa = 11.84WILPTLKK10 pKa = 10.29LARR13 pKa = 11.84VRR15 pKa = 11.84TLPVVLNKK23 pKa = 9.78IKK25 pKa = 9.81KK26 pKa = 8.41TITSFKK32 pKa = 10.65KK33 pKa = 10.45SLQQLSTNSPQRR45 pKa = 11.84MFRR48 pKa = 11.84SSLKK52 pKa = 10.44KK53 pKa = 10.38
MM1 pKa = 7.31KK2 pKa = 10.19RR3 pKa = 11.84WILPTLKK10 pKa = 10.29LARR13 pKa = 11.84VRR15 pKa = 11.84TLPVVLNKK23 pKa = 9.78IKK25 pKa = 9.81KK26 pKa = 8.41TITSFKK32 pKa = 10.65KK33 pKa = 10.45SLQQLSTNSPQRR45 pKa = 11.84MFRR48 pKa = 11.84SSLKK52 pKa = 10.44KK53 pKa = 10.38
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9185206 |
24 |
10474 |
392.6 |
43.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.859 ± 0.015 | 1.621 ± 0.008 |
5.698 ± 0.011 | 6.655 ± 0.023 |
3.711 ± 0.011 | 6.046 ± 0.019 |
2.315 ± 0.008 | 4.14 ± 0.012 |
5.168 ± 0.017 | 9.335 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.477 ± 0.008 | 3.582 ± 0.009 |
4.7 ± 0.014 | 4.226 ± 0.014 |
6.22 ± 0.02 | 8.289 ± 0.021 |
6.018 ± 0.018 | 7.121 ± 0.016 |
1.209 ± 0.005 | 2.604 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
