
Maribacter sp. RZ26
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter; unclassified Maribacter
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
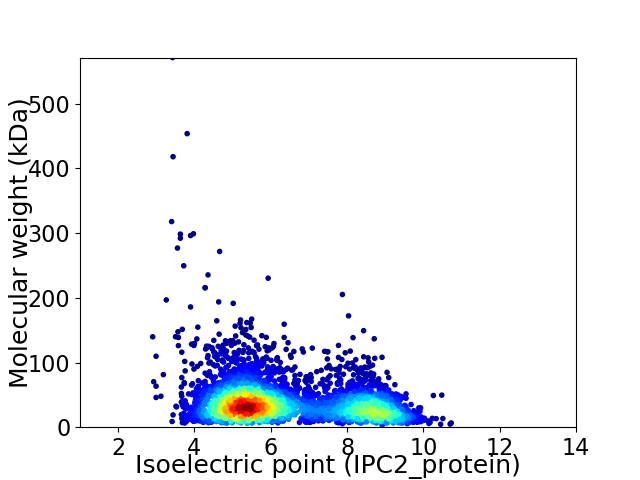
Virtual 2D-PAGE plot for 4327 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S3PTN6|A0A5S3PTN6_9FLAO Uncharacterized protein OS=Maribacter sp. RZ26 OX=2578118 GN=FEE95_12415 PE=4 SV=1
MM1 pKa = 7.81NSIQPSKK8 pKa = 10.58FPQNNMDD15 pKa = 4.86RR16 pKa = 11.84ILLYY20 pKa = 10.79FMLMASFMATSQTTFIEE37 pKa = 4.45SATAHH42 pKa = 6.34GLNISGNKK50 pKa = 9.86DD51 pKa = 3.09GGHH54 pKa = 6.54AWSDD58 pKa = 3.42FDD60 pKa = 5.89RR61 pKa = 11.84DD62 pKa = 2.99GDD64 pKa = 4.17LDD66 pKa = 4.01VLVLEE71 pKa = 4.56NTNSKK76 pKa = 10.63SYY78 pKa = 10.99LMRR81 pKa = 11.84NNSGTYY87 pKa = 10.55SNVQATLAPGMLSSAAEE104 pKa = 4.04RR105 pKa = 11.84QAVWGDD111 pKa = 3.47LNNDD115 pKa = 3.15GRR117 pKa = 11.84PDD119 pKa = 3.7FMIGSHH125 pKa = 6.5GNTNSNPAFQIFIQNQDD142 pKa = 2.64GSFGDD147 pKa = 3.76GKK149 pKa = 10.97GGYY152 pKa = 7.59TPITVGRR159 pKa = 11.84SGYY162 pKa = 8.69TININPINAEE172 pKa = 3.71GAGFFDD178 pKa = 5.29FEE180 pKa = 5.45GDD182 pKa = 3.18GDD184 pKa = 4.44LDD186 pKa = 4.12IFFDD190 pKa = 3.52SHH192 pKa = 7.59NYY194 pKa = 9.96GIEE197 pKa = 4.0LLRR200 pKa = 11.84NNYY203 pKa = 9.03ISHH206 pKa = 7.02ISHH209 pKa = 7.43TIINPPATSMFTHH222 pKa = 6.57ITTGNGNGVVNFGLNQFASDD242 pKa = 3.7GDD244 pKa = 4.09YY245 pKa = 10.01GTAADD250 pKa = 4.3VDD252 pKa = 3.97DD253 pKa = 5.88DD254 pKa = 4.05GWVDD258 pKa = 2.52IFMRR262 pKa = 11.84KK263 pKa = 9.45RR264 pKa = 11.84NEE266 pKa = 3.49NDD268 pKa = 2.7FFLNQGGVFSNGSDD282 pKa = 3.67LAQAHH287 pKa = 6.06NNNKK291 pKa = 9.54GGNGLWDD298 pKa = 4.35LDD300 pKa = 3.81NDD302 pKa = 4.14GDD304 pKa = 5.06LDD306 pKa = 4.76AVWTEE311 pKa = 3.7NDD313 pKa = 2.88LTQIFRR319 pKa = 11.84NDD321 pKa = 3.46GPGVWTALGATVFPGLPQPSNTNQGNSNASIDD353 pKa = 3.76ALAGGDD359 pKa = 3.72IDD361 pKa = 4.66NDD363 pKa = 3.57GDD365 pKa = 3.55IDD367 pKa = 4.88IILVGNNRR375 pKa = 11.84SYY377 pKa = 11.66LYY379 pKa = 10.2INQLNSPTPAPGVIGSGAAMTFSLDD404 pKa = 3.34PQTFNSSNGEE414 pKa = 3.95GTTMVDD420 pKa = 4.35FDD422 pKa = 6.3DD423 pKa = 6.71DD424 pKa = 4.61GDD426 pKa = 4.26LDD428 pKa = 3.81IYY430 pKa = 10.78MSISGTNRR438 pKa = 11.84LYY440 pKa = 11.34VNQLPVANRR449 pKa = 11.84NNHH452 pKa = 5.36LMVNVSEE459 pKa = 4.89DD460 pKa = 3.45RR461 pKa = 11.84NADD464 pKa = 3.19GSTGGFAGRR473 pKa = 11.84TAIGTNVLIRR483 pKa = 11.84DD484 pKa = 3.77CDD486 pKa = 3.81GNIISGLRR494 pKa = 11.84QVNGVFGHH502 pKa = 6.3GTQQPEE508 pKa = 4.14EE509 pKa = 4.14VHH511 pKa = 6.65FGLPLGEE518 pKa = 4.06NQTYY522 pKa = 10.21LIEE525 pKa = 3.88VHH527 pKa = 6.14YY528 pKa = 10.35PNFYY532 pKa = 11.15DD533 pKa = 5.26PIDD536 pKa = 3.8GFTRR540 pKa = 11.84LIATGIAQPSLIAGTNHH557 pKa = 5.14YY558 pKa = 9.17TLTTTDD564 pKa = 4.45AEE566 pKa = 4.43SLQNEE571 pKa = 4.45NAPVANDD578 pKa = 3.45DD579 pKa = 4.36FEE581 pKa = 4.66TVSYY585 pKa = 8.26GTAVSVQINLFDD597 pKa = 4.66NDD599 pKa = 4.32SEE601 pKa = 4.49PDD603 pKa = 3.45GEE605 pKa = 4.22NFYY608 pKa = 10.64IEE610 pKa = 5.71SIVQPAIGSVTIDD623 pKa = 3.6DD624 pKa = 4.53ADD626 pKa = 3.94NGLVTYY632 pKa = 8.32TYY634 pKa = 10.82SAGIPFPGATFDD646 pKa = 3.86YY647 pKa = 9.52TISDD651 pKa = 3.9AQVTLCPSAGLSDD664 pKa = 3.96SATVTIDD671 pKa = 3.85EE672 pKa = 4.82PCADD676 pKa = 3.9PTGVDD681 pKa = 3.34TDD683 pKa = 3.79GDD685 pKa = 4.56GINDD689 pKa = 3.63VCDD692 pKa = 4.85LDD694 pKa = 5.46DD695 pKa = 6.05DD696 pKa = 4.6NDD698 pKa = 5.07GILDD702 pKa = 3.67TVEE705 pKa = 3.84RR706 pKa = 11.84AKK708 pKa = 10.23TVLWVTQGTPEE719 pKa = 4.1TEE721 pKa = 4.09EE722 pKa = 3.76QNTIDD727 pKa = 4.44KK728 pKa = 8.77LTALGYY734 pKa = 8.66TVTVVDD740 pKa = 5.13DD741 pKa = 4.16NVGGNANDD749 pKa = 3.82FSVTFLYY756 pKa = 10.94EE757 pKa = 4.11DD758 pKa = 4.03VNSGTAYY765 pKa = 10.73ANVANLSTTTNGVITSEE782 pKa = 3.77NALFDD787 pKa = 5.14EE788 pKa = 4.69ILGTSGATGNTNTNFINITDD808 pKa = 3.81NTHH811 pKa = 7.44PITSNLPLGNYY822 pKa = 9.69DD823 pKa = 3.29VGDD826 pKa = 3.35AAFYY830 pKa = 10.44VNNVVSGQKK839 pKa = 10.21LGNHH843 pKa = 6.97PNGQAVLIVWEE854 pKa = 5.09EE855 pKa = 3.8GDD857 pKa = 4.97AMDD860 pKa = 4.82TGIAPGRR867 pKa = 11.84RR868 pKa = 11.84TVLPLTNSTGGFNSNGEE885 pKa = 4.24DD886 pKa = 3.6LLVNAIIWTGNIDD899 pKa = 3.49TDD901 pKa = 3.44EE902 pKa = 5.67DD903 pKa = 4.61GIADD907 pKa = 4.3HH908 pKa = 7.25LDD910 pKa = 3.45SDD912 pKa = 5.08SDD914 pKa = 4.51ADD916 pKa = 3.87GCNDD920 pKa = 4.62AIEE923 pKa = 4.95AYY925 pKa = 10.28SNLSADD931 pKa = 4.13PDD933 pKa = 4.24SNDD936 pKa = 3.27YY937 pKa = 11.13YY938 pKa = 11.14GTGNPPPVNTDD949 pKa = 3.09GTVVAASYY957 pKa = 7.51QTPADD962 pKa = 3.55NDD964 pKa = 4.22SNGTYY969 pKa = 10.58DD970 pKa = 3.52FQEE973 pKa = 4.7AGTAPSISSQPANLTICPGCSGTFSVTASNSDD1005 pKa = 3.32GYY1007 pKa = 10.57QWQTFNGTSWVDD1019 pKa = 3.57LTDD1022 pKa = 3.63SGIHH1026 pKa = 6.24SGTTTSTLNITNATPSDD1043 pKa = 3.43NGNQYY1048 pKa = 10.47RR1049 pKa = 11.84VLVSNSLFICTTIISNTTILTVNVSSVITNRR1080 pKa = 11.84RR1081 pKa = 11.84ITYY1084 pKa = 8.77RR1085 pKa = 11.84VKK1087 pKa = 10.43MNN1089 pKa = 3.57
MM1 pKa = 7.81NSIQPSKK8 pKa = 10.58FPQNNMDD15 pKa = 4.86RR16 pKa = 11.84ILLYY20 pKa = 10.79FMLMASFMATSQTTFIEE37 pKa = 4.45SATAHH42 pKa = 6.34GLNISGNKK50 pKa = 9.86DD51 pKa = 3.09GGHH54 pKa = 6.54AWSDD58 pKa = 3.42FDD60 pKa = 5.89RR61 pKa = 11.84DD62 pKa = 2.99GDD64 pKa = 4.17LDD66 pKa = 4.01VLVLEE71 pKa = 4.56NTNSKK76 pKa = 10.63SYY78 pKa = 10.99LMRR81 pKa = 11.84NNSGTYY87 pKa = 10.55SNVQATLAPGMLSSAAEE104 pKa = 4.04RR105 pKa = 11.84QAVWGDD111 pKa = 3.47LNNDD115 pKa = 3.15GRR117 pKa = 11.84PDD119 pKa = 3.7FMIGSHH125 pKa = 6.5GNTNSNPAFQIFIQNQDD142 pKa = 2.64GSFGDD147 pKa = 3.76GKK149 pKa = 10.97GGYY152 pKa = 7.59TPITVGRR159 pKa = 11.84SGYY162 pKa = 8.69TININPINAEE172 pKa = 3.71GAGFFDD178 pKa = 5.29FEE180 pKa = 5.45GDD182 pKa = 3.18GDD184 pKa = 4.44LDD186 pKa = 4.12IFFDD190 pKa = 3.52SHH192 pKa = 7.59NYY194 pKa = 9.96GIEE197 pKa = 4.0LLRR200 pKa = 11.84NNYY203 pKa = 9.03ISHH206 pKa = 7.02ISHH209 pKa = 7.43TIINPPATSMFTHH222 pKa = 6.57ITTGNGNGVVNFGLNQFASDD242 pKa = 3.7GDD244 pKa = 4.09YY245 pKa = 10.01GTAADD250 pKa = 4.3VDD252 pKa = 3.97DD253 pKa = 5.88DD254 pKa = 4.05GWVDD258 pKa = 2.52IFMRR262 pKa = 11.84KK263 pKa = 9.45RR264 pKa = 11.84NEE266 pKa = 3.49NDD268 pKa = 2.7FFLNQGGVFSNGSDD282 pKa = 3.67LAQAHH287 pKa = 6.06NNNKK291 pKa = 9.54GGNGLWDD298 pKa = 4.35LDD300 pKa = 3.81NDD302 pKa = 4.14GDD304 pKa = 5.06LDD306 pKa = 4.76AVWTEE311 pKa = 3.7NDD313 pKa = 2.88LTQIFRR319 pKa = 11.84NDD321 pKa = 3.46GPGVWTALGATVFPGLPQPSNTNQGNSNASIDD353 pKa = 3.76ALAGGDD359 pKa = 3.72IDD361 pKa = 4.66NDD363 pKa = 3.57GDD365 pKa = 3.55IDD367 pKa = 4.88IILVGNNRR375 pKa = 11.84SYY377 pKa = 11.66LYY379 pKa = 10.2INQLNSPTPAPGVIGSGAAMTFSLDD404 pKa = 3.34PQTFNSSNGEE414 pKa = 3.95GTTMVDD420 pKa = 4.35FDD422 pKa = 6.3DD423 pKa = 6.71DD424 pKa = 4.61GDD426 pKa = 4.26LDD428 pKa = 3.81IYY430 pKa = 10.78MSISGTNRR438 pKa = 11.84LYY440 pKa = 11.34VNQLPVANRR449 pKa = 11.84NNHH452 pKa = 5.36LMVNVSEE459 pKa = 4.89DD460 pKa = 3.45RR461 pKa = 11.84NADD464 pKa = 3.19GSTGGFAGRR473 pKa = 11.84TAIGTNVLIRR483 pKa = 11.84DD484 pKa = 3.77CDD486 pKa = 3.81GNIISGLRR494 pKa = 11.84QVNGVFGHH502 pKa = 6.3GTQQPEE508 pKa = 4.14EE509 pKa = 4.14VHH511 pKa = 6.65FGLPLGEE518 pKa = 4.06NQTYY522 pKa = 10.21LIEE525 pKa = 3.88VHH527 pKa = 6.14YY528 pKa = 10.35PNFYY532 pKa = 11.15DD533 pKa = 5.26PIDD536 pKa = 3.8GFTRR540 pKa = 11.84LIATGIAQPSLIAGTNHH557 pKa = 5.14YY558 pKa = 9.17TLTTTDD564 pKa = 4.45AEE566 pKa = 4.43SLQNEE571 pKa = 4.45NAPVANDD578 pKa = 3.45DD579 pKa = 4.36FEE581 pKa = 4.66TVSYY585 pKa = 8.26GTAVSVQINLFDD597 pKa = 4.66NDD599 pKa = 4.32SEE601 pKa = 4.49PDD603 pKa = 3.45GEE605 pKa = 4.22NFYY608 pKa = 10.64IEE610 pKa = 5.71SIVQPAIGSVTIDD623 pKa = 3.6DD624 pKa = 4.53ADD626 pKa = 3.94NGLVTYY632 pKa = 8.32TYY634 pKa = 10.82SAGIPFPGATFDD646 pKa = 3.86YY647 pKa = 9.52TISDD651 pKa = 3.9AQVTLCPSAGLSDD664 pKa = 3.96SATVTIDD671 pKa = 3.85EE672 pKa = 4.82PCADD676 pKa = 3.9PTGVDD681 pKa = 3.34TDD683 pKa = 3.79GDD685 pKa = 4.56GINDD689 pKa = 3.63VCDD692 pKa = 4.85LDD694 pKa = 5.46DD695 pKa = 6.05DD696 pKa = 4.6NDD698 pKa = 5.07GILDD702 pKa = 3.67TVEE705 pKa = 3.84RR706 pKa = 11.84AKK708 pKa = 10.23TVLWVTQGTPEE719 pKa = 4.1TEE721 pKa = 4.09EE722 pKa = 3.76QNTIDD727 pKa = 4.44KK728 pKa = 8.77LTALGYY734 pKa = 8.66TVTVVDD740 pKa = 5.13DD741 pKa = 4.16NVGGNANDD749 pKa = 3.82FSVTFLYY756 pKa = 10.94EE757 pKa = 4.11DD758 pKa = 4.03VNSGTAYY765 pKa = 10.73ANVANLSTTTNGVITSEE782 pKa = 3.77NALFDD787 pKa = 5.14EE788 pKa = 4.69ILGTSGATGNTNTNFINITDD808 pKa = 3.81NTHH811 pKa = 7.44PITSNLPLGNYY822 pKa = 9.69DD823 pKa = 3.29VGDD826 pKa = 3.35AAFYY830 pKa = 10.44VNNVVSGQKK839 pKa = 10.21LGNHH843 pKa = 6.97PNGQAVLIVWEE854 pKa = 5.09EE855 pKa = 3.8GDD857 pKa = 4.97AMDD860 pKa = 4.82TGIAPGRR867 pKa = 11.84RR868 pKa = 11.84TVLPLTNSTGGFNSNGEE885 pKa = 4.24DD886 pKa = 3.6LLVNAIIWTGNIDD899 pKa = 3.49TDD901 pKa = 3.44EE902 pKa = 5.67DD903 pKa = 4.61GIADD907 pKa = 4.3HH908 pKa = 7.25LDD910 pKa = 3.45SDD912 pKa = 5.08SDD914 pKa = 4.51ADD916 pKa = 3.87GCNDD920 pKa = 4.62AIEE923 pKa = 4.95AYY925 pKa = 10.28SNLSADD931 pKa = 4.13PDD933 pKa = 4.24SNDD936 pKa = 3.27YY937 pKa = 11.13YY938 pKa = 11.14GTGNPPPVNTDD949 pKa = 3.09GTVVAASYY957 pKa = 7.51QTPADD962 pKa = 3.55NDD964 pKa = 4.22SNGTYY969 pKa = 10.58DD970 pKa = 3.52FQEE973 pKa = 4.7AGTAPSISSQPANLTICPGCSGTFSVTASNSDD1005 pKa = 3.32GYY1007 pKa = 10.57QWQTFNGTSWVDD1019 pKa = 3.57LTDD1022 pKa = 3.63SGIHH1026 pKa = 6.24SGTTTSTLNITNATPSDD1043 pKa = 3.43NGNQYY1048 pKa = 10.47RR1049 pKa = 11.84VLVSNSLFICTTIISNTTILTVNVSSVITNRR1080 pKa = 11.84RR1081 pKa = 11.84ITYY1084 pKa = 8.77RR1085 pKa = 11.84VKK1087 pKa = 10.43MNN1089 pKa = 3.57
Molecular weight: 115.95 kDa
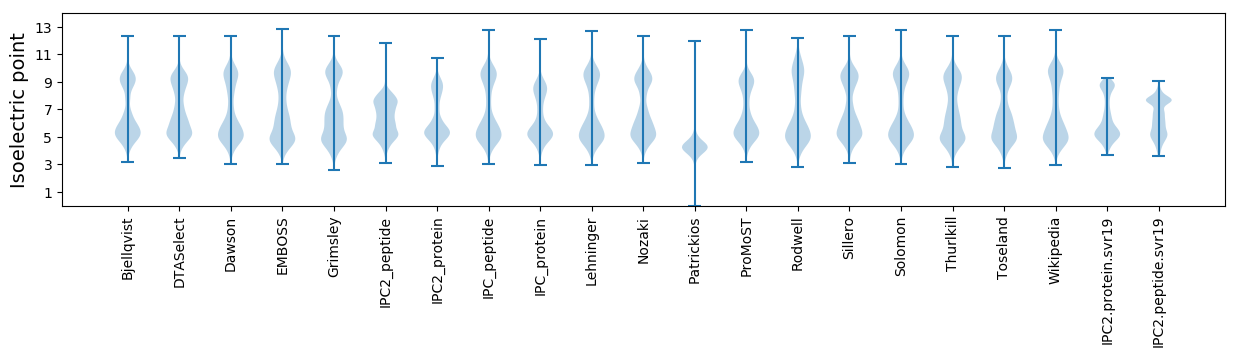
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S3PVC9|A0A5S3PVC9_9FLAO Crp/Fnr family transcriptional regulator OS=Maribacter sp. RZ26 OX=2578118 GN=FEE95_05685 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.57SLLAPLKK38 pKa = 10.66KK39 pKa = 10.41SGGRR43 pKa = 11.84NSQGKK48 pKa = 5.92MTIRR52 pKa = 11.84QKK54 pKa = 11.2GGGHH58 pKa = 5.43KK59 pKa = 9.79RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84VIDD66 pKa = 4.1FKK68 pKa = 11.07RR69 pKa = 11.84DD70 pKa = 3.59KK71 pKa = 10.93QDD73 pKa = 2.67VTATVKK79 pKa = 10.61SIQYY83 pKa = 9.95DD84 pKa = 3.58PNRR87 pKa = 11.84TAFIALLEE95 pKa = 4.24YY96 pKa = 10.92VDD98 pKa = 3.68GEE100 pKa = 3.98KK101 pKa = 10.33RR102 pKa = 11.84YY103 pKa = 10.25IIAQNGLQVDD113 pKa = 3.85QKK115 pKa = 9.61VTSGANAAPEE125 pKa = 4.12IGNALPLSAIPLGTIISCIEE145 pKa = 3.75LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 9.93DD168 pKa = 4.08GKK170 pKa = 10.42FVTVKK175 pKa = 9.72MPSGEE180 pKa = 4.12TRR182 pKa = 11.84LILSDD187 pKa = 3.82CLATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.27GYY255 pKa = 7.48RR256 pKa = 11.84TRR258 pKa = 11.84QKK260 pKa = 9.45TKK262 pKa = 9.43STNKK266 pKa = 10.22YY267 pKa = 8.44ILEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.2KK274 pKa = 9.94
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.57SLLAPLKK38 pKa = 10.66KK39 pKa = 10.41SGGRR43 pKa = 11.84NSQGKK48 pKa = 5.92MTIRR52 pKa = 11.84QKK54 pKa = 11.2GGGHH58 pKa = 5.43KK59 pKa = 9.79RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84VIDD66 pKa = 4.1FKK68 pKa = 11.07RR69 pKa = 11.84DD70 pKa = 3.59KK71 pKa = 10.93QDD73 pKa = 2.67VTATVKK79 pKa = 10.61SIQYY83 pKa = 9.95DD84 pKa = 3.58PNRR87 pKa = 11.84TAFIALLEE95 pKa = 4.24YY96 pKa = 10.92VDD98 pKa = 3.68GEE100 pKa = 3.98KK101 pKa = 10.33RR102 pKa = 11.84YY103 pKa = 10.25IIAQNGLQVDD113 pKa = 3.85QKK115 pKa = 9.61VTSGANAAPEE125 pKa = 4.12IGNALPLSAIPLGTIISCIEE145 pKa = 3.75LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 9.93DD168 pKa = 4.08GKK170 pKa = 10.42FVTVKK175 pKa = 9.72MPSGEE180 pKa = 4.12TRR182 pKa = 11.84LILSDD187 pKa = 3.82CLATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.27GYY255 pKa = 7.48RR256 pKa = 11.84TRR258 pKa = 11.84QKK260 pKa = 9.45TKK262 pKa = 9.43STNKK266 pKa = 10.22YY267 pKa = 8.44ILEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.2KK274 pKa = 9.94
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1542337 |
30 |
5436 |
356.4 |
40.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.471 ± 0.036 | 0.752 ± 0.015 |
5.805 ± 0.035 | 6.811 ± 0.039 |
5.18 ± 0.029 | 6.855 ± 0.043 |
1.765 ± 0.019 | 7.441 ± 0.037 |
7.37 ± 0.056 | 9.293 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.018 | 5.772 ± 0.039 |
3.58 ± 0.023 | 3.296 ± 0.019 |
3.6 ± 0.023 | 6.592 ± 0.033 |
5.76 ± 0.052 | 6.269 ± 0.029 |
1.191 ± 0.016 | 3.919 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
