
Candidatus Accumulibacter sp. SK-02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Accumulibacter; unclassified Candidatus Accumulibacter
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
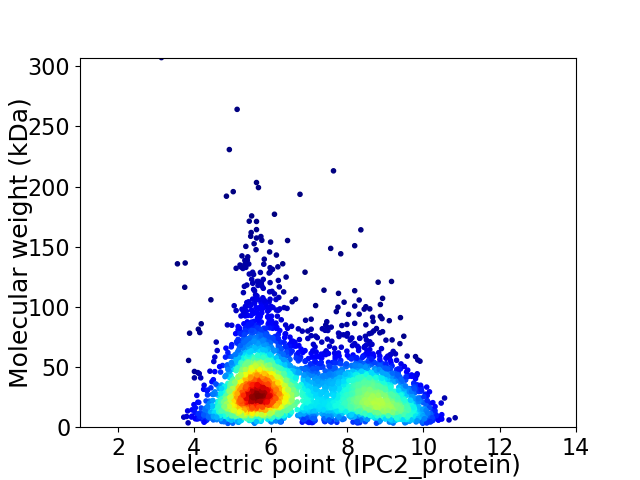
Virtual 2D-PAGE plot for 3953 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A080M8W4|A0A080M8W4_9PROT Uncharacterized protein OS=Candidatus Accumulibacter sp. SK-02 OX=1453999 GN=AW06_002035 PE=4 SV=1
MM1 pKa = 7.6ALASWTQQQILAQLDD16 pKa = 4.22SGSHH20 pKa = 4.6WSGSTVTYY28 pKa = 10.5AFPTSSSGLYY38 pKa = 10.31GSQEE42 pKa = 3.69QAGFQGLNSVQQAAATLALQTWDD65 pKa = 4.98DD66 pKa = 5.47LITPDD71 pKa = 4.11LAKK74 pKa = 9.98VTSGSSNIEE83 pKa = 3.93FGTSSTGVEE92 pKa = 3.98YY93 pKa = 11.03AHH95 pKa = 7.37AYY97 pKa = 9.84FPTTGSVWFSRR108 pKa = 11.84AYY110 pKa = 10.42SDD112 pKa = 5.03LMNPQVGAHH121 pKa = 6.04SFLSYY126 pKa = 9.9VHH128 pKa = 6.97EE129 pKa = 4.97IGHH132 pKa = 6.83AFGLDD137 pKa = 3.1HH138 pKa = 6.75MGDD141 pKa = 4.0YY142 pKa = 11.11NGGGSWTPSSYY153 pKa = 10.58QDD155 pKa = 3.11SGVYY159 pKa = 10.31SVMSYY164 pKa = 10.11FGPNWGSGSSSGVGLVAWADD184 pKa = 3.38WVGADD189 pKa = 3.8GKK191 pKa = 10.57LYY193 pKa = 10.79SPQTPMLNDD202 pKa = 3.25IMAMQSMYY210 pKa = 10.77GAEE213 pKa = 4.17TTTRR217 pKa = 11.84TEE219 pKa = 3.69DD220 pKa = 3.0TTYY223 pKa = 11.34GFGSSDD229 pKa = 3.72FGTLSAIYY237 pKa = 9.46NFSVNLHH244 pKa = 7.08PILTIYY250 pKa = 10.78DD251 pKa = 3.71SAGTDD256 pKa = 3.54TLNLSGWSTPSLIDD270 pKa = 3.96LAPGAYY276 pKa = 9.65SSCNSMTYY284 pKa = 10.46NIAIAYY290 pKa = 6.18TCNIEE295 pKa = 4.0NAIGGAGADD304 pKa = 3.83TITGNSLNNYY314 pKa = 9.45LDD316 pKa = 3.99GGSGNDD322 pKa = 3.71SLTGGAGDD330 pKa = 3.87DD331 pKa = 3.92TLVAGAGNDD340 pKa = 3.62ALDD343 pKa = 4.08GGGGNDD349 pKa = 3.02TVVFAGTFSEE359 pKa = 4.88YY360 pKa = 10.31TFSYY364 pKa = 9.99TINQGFTFASSGSGSDD380 pKa = 3.48LIKK383 pKa = 10.89NVEE386 pKa = 4.11TFVFSNVTKK395 pKa = 10.61SASEE399 pKa = 4.13LSADD403 pKa = 4.02ASSDD407 pKa = 3.65PDD409 pKa = 3.85PGSVQVSIAAGAASIAEE426 pKa = 4.41GNSGSTAYY434 pKa = 10.39VFNVSLSKK442 pKa = 10.9ASEE445 pKa = 4.15TTQTVDD451 pKa = 2.39WRR453 pKa = 11.84LDD455 pKa = 3.41GSGASPATSSDD466 pKa = 3.36FTGATSGVVTFQPGEE481 pKa = 4.1TTKK484 pKa = 10.24TIQVLVVGDD493 pKa = 3.79TVVEE497 pKa = 4.07SDD499 pKa = 3.72EE500 pKa = 4.83GFTLSLSNPSSGLVLGTASAAGTIRR525 pKa = 11.84NDD527 pKa = 3.61DD528 pKa = 3.61VTAVPDD534 pKa = 4.69DD535 pKa = 3.97YY536 pKa = 11.84PMTTNTRR543 pKa = 11.84GAITVGGAAVSGVIEE558 pKa = 4.11QASDD562 pKa = 3.24GDD564 pKa = 4.2LFKK567 pKa = 11.02VTLKK571 pKa = 11.03ANNSYY576 pKa = 10.14IFKK579 pKa = 9.95LARR582 pKa = 11.84VSGSLDD588 pKa = 3.41PYY590 pKa = 11.28LEE592 pKa = 4.86LYY594 pKa = 10.17SSKK597 pKa = 10.65LAVITQNDD605 pKa = 3.58NAGLSTTDD613 pKa = 2.93SQIIYY618 pKa = 8.98TPTAGGTYY626 pKa = 10.35YY627 pKa = 11.05LAAKK631 pKa = 10.2DD632 pKa = 3.99RR633 pKa = 11.84AQSTGSYY640 pKa = 9.73LISAVPCSGLTIEE653 pKa = 6.01GDD655 pKa = 3.45DD656 pKa = 3.79TVNVLTGTAGNDD668 pKa = 3.17SLYY671 pKa = 11.28GLGGNDD677 pKa = 3.37TLNAGMGNDD686 pKa = 4.4LLDD689 pKa = 4.41GGAGMDD695 pKa = 3.55QLAGGKK701 pKa = 10.19GDD703 pKa = 5.79DD704 pKa = 4.08IYY706 pKa = 11.63LVDD709 pKa = 3.78NVSDD713 pKa = 3.91VVIEE717 pKa = 4.15SSAADD722 pKa = 3.41GKK724 pKa = 10.81DD725 pKa = 3.25LVKK728 pKa = 11.04ASVSWTLGANVEE740 pKa = 4.23KK741 pKa = 11.27LEE743 pKa = 4.19LTGTANLMGTGNTLANIITGNSGNNVLNGLGGVDD777 pKa = 3.73SLNGGEE783 pKa = 5.15GSDD786 pKa = 4.15LYY788 pKa = 11.55LFGLPAEE795 pKa = 4.62HH796 pKa = 6.99SSAEE800 pKa = 4.03IADD803 pKa = 3.94TGSTGVDD810 pKa = 3.24EE811 pKa = 4.9VRR813 pKa = 11.84FTATGAGTLKK823 pKa = 10.64LYY825 pKa = 10.96AGDD828 pKa = 3.64TGIEE832 pKa = 3.83RR833 pKa = 11.84VVIGTGTDD841 pKa = 3.05AVANSEE847 pKa = 4.17GTVALNVDD855 pKa = 4.4ASPLSNAITMIGNAGANILTGTAFADD881 pKa = 3.7QMDD884 pKa = 4.52GGSGNDD890 pKa = 4.11LLNGQAGIDD899 pKa = 3.77LMDD902 pKa = 4.36GEE904 pKa = 4.99EE905 pKa = 4.51GSDD908 pKa = 2.93IYY910 pKa = 11.28LIGLAAAHH918 pKa = 6.0TAAEE922 pKa = 4.24IADD925 pKa = 4.01TGSSGVDD932 pKa = 3.19EE933 pKa = 4.6VRR935 pKa = 11.84FAATTASTLTLFAGDD950 pKa = 3.53TGIEE954 pKa = 3.89RR955 pKa = 11.84VVLGTGTGAVATSTGSAALNADD977 pKa = 3.18ASALVNAITLVGNAGANILTGTEE1000 pKa = 3.59FADD1003 pKa = 4.89RR1004 pKa = 11.84IEE1006 pKa = 4.72GGGGADD1012 pKa = 3.3QLFGGSGNDD1021 pKa = 3.48TLFGGLGNDD1030 pKa = 3.48TLTGGEE1036 pKa = 4.41GEE1038 pKa = 4.39DD1039 pKa = 3.34HH1040 pKa = 6.82FVLNTAANAANNRR1053 pKa = 11.84DD1054 pKa = 3.51VLVDD1058 pKa = 4.1FEE1060 pKa = 4.93SGADD1064 pKa = 3.73HH1065 pKa = 7.33IEE1067 pKa = 3.98LSLLVFKK1074 pKa = 11.11GLGKK1078 pKa = 7.36TTGVLGEE1085 pKa = 4.06AQFWAAADD1093 pKa = 3.61AVKK1096 pKa = 10.64GHH1098 pKa = 7.01DD1099 pKa = 3.36ADD1101 pKa = 5.4DD1102 pKa = 3.46RR1103 pKa = 11.84VIYY1106 pKa = 8.32NTSTGSLYY1114 pKa = 10.94YY1115 pKa = 10.66DD1116 pKa = 3.89ADD1118 pKa = 3.97GSGKK1122 pKa = 10.06GAAVEE1127 pKa = 4.0IAVLGLSTHH1136 pKa = 6.56PTLEE1140 pKa = 4.51HH1141 pKa = 7.09GDD1143 pKa = 3.72FLLII1147 pKa = 4.64
MM1 pKa = 7.6ALASWTQQQILAQLDD16 pKa = 4.22SGSHH20 pKa = 4.6WSGSTVTYY28 pKa = 10.5AFPTSSSGLYY38 pKa = 10.31GSQEE42 pKa = 3.69QAGFQGLNSVQQAAATLALQTWDD65 pKa = 4.98DD66 pKa = 5.47LITPDD71 pKa = 4.11LAKK74 pKa = 9.98VTSGSSNIEE83 pKa = 3.93FGTSSTGVEE92 pKa = 3.98YY93 pKa = 11.03AHH95 pKa = 7.37AYY97 pKa = 9.84FPTTGSVWFSRR108 pKa = 11.84AYY110 pKa = 10.42SDD112 pKa = 5.03LMNPQVGAHH121 pKa = 6.04SFLSYY126 pKa = 9.9VHH128 pKa = 6.97EE129 pKa = 4.97IGHH132 pKa = 6.83AFGLDD137 pKa = 3.1HH138 pKa = 6.75MGDD141 pKa = 4.0YY142 pKa = 11.11NGGGSWTPSSYY153 pKa = 10.58QDD155 pKa = 3.11SGVYY159 pKa = 10.31SVMSYY164 pKa = 10.11FGPNWGSGSSSGVGLVAWADD184 pKa = 3.38WVGADD189 pKa = 3.8GKK191 pKa = 10.57LYY193 pKa = 10.79SPQTPMLNDD202 pKa = 3.25IMAMQSMYY210 pKa = 10.77GAEE213 pKa = 4.17TTTRR217 pKa = 11.84TEE219 pKa = 3.69DD220 pKa = 3.0TTYY223 pKa = 11.34GFGSSDD229 pKa = 3.72FGTLSAIYY237 pKa = 9.46NFSVNLHH244 pKa = 7.08PILTIYY250 pKa = 10.78DD251 pKa = 3.71SAGTDD256 pKa = 3.54TLNLSGWSTPSLIDD270 pKa = 3.96LAPGAYY276 pKa = 9.65SSCNSMTYY284 pKa = 10.46NIAIAYY290 pKa = 6.18TCNIEE295 pKa = 4.0NAIGGAGADD304 pKa = 3.83TITGNSLNNYY314 pKa = 9.45LDD316 pKa = 3.99GGSGNDD322 pKa = 3.71SLTGGAGDD330 pKa = 3.87DD331 pKa = 3.92TLVAGAGNDD340 pKa = 3.62ALDD343 pKa = 4.08GGGGNDD349 pKa = 3.02TVVFAGTFSEE359 pKa = 4.88YY360 pKa = 10.31TFSYY364 pKa = 9.99TINQGFTFASSGSGSDD380 pKa = 3.48LIKK383 pKa = 10.89NVEE386 pKa = 4.11TFVFSNVTKK395 pKa = 10.61SASEE399 pKa = 4.13LSADD403 pKa = 4.02ASSDD407 pKa = 3.65PDD409 pKa = 3.85PGSVQVSIAAGAASIAEE426 pKa = 4.41GNSGSTAYY434 pKa = 10.39VFNVSLSKK442 pKa = 10.9ASEE445 pKa = 4.15TTQTVDD451 pKa = 2.39WRR453 pKa = 11.84LDD455 pKa = 3.41GSGASPATSSDD466 pKa = 3.36FTGATSGVVTFQPGEE481 pKa = 4.1TTKK484 pKa = 10.24TIQVLVVGDD493 pKa = 3.79TVVEE497 pKa = 4.07SDD499 pKa = 3.72EE500 pKa = 4.83GFTLSLSNPSSGLVLGTASAAGTIRR525 pKa = 11.84NDD527 pKa = 3.61DD528 pKa = 3.61VTAVPDD534 pKa = 4.69DD535 pKa = 3.97YY536 pKa = 11.84PMTTNTRR543 pKa = 11.84GAITVGGAAVSGVIEE558 pKa = 4.11QASDD562 pKa = 3.24GDD564 pKa = 4.2LFKK567 pKa = 11.02VTLKK571 pKa = 11.03ANNSYY576 pKa = 10.14IFKK579 pKa = 9.95LARR582 pKa = 11.84VSGSLDD588 pKa = 3.41PYY590 pKa = 11.28LEE592 pKa = 4.86LYY594 pKa = 10.17SSKK597 pKa = 10.65LAVITQNDD605 pKa = 3.58NAGLSTTDD613 pKa = 2.93SQIIYY618 pKa = 8.98TPTAGGTYY626 pKa = 10.35YY627 pKa = 11.05LAAKK631 pKa = 10.2DD632 pKa = 3.99RR633 pKa = 11.84AQSTGSYY640 pKa = 9.73LISAVPCSGLTIEE653 pKa = 6.01GDD655 pKa = 3.45DD656 pKa = 3.79TVNVLTGTAGNDD668 pKa = 3.17SLYY671 pKa = 11.28GLGGNDD677 pKa = 3.37TLNAGMGNDD686 pKa = 4.4LLDD689 pKa = 4.41GGAGMDD695 pKa = 3.55QLAGGKK701 pKa = 10.19GDD703 pKa = 5.79DD704 pKa = 4.08IYY706 pKa = 11.63LVDD709 pKa = 3.78NVSDD713 pKa = 3.91VVIEE717 pKa = 4.15SSAADD722 pKa = 3.41GKK724 pKa = 10.81DD725 pKa = 3.25LVKK728 pKa = 11.04ASVSWTLGANVEE740 pKa = 4.23KK741 pKa = 11.27LEE743 pKa = 4.19LTGTANLMGTGNTLANIITGNSGNNVLNGLGGVDD777 pKa = 3.73SLNGGEE783 pKa = 5.15GSDD786 pKa = 4.15LYY788 pKa = 11.55LFGLPAEE795 pKa = 4.62HH796 pKa = 6.99SSAEE800 pKa = 4.03IADD803 pKa = 3.94TGSTGVDD810 pKa = 3.24EE811 pKa = 4.9VRR813 pKa = 11.84FTATGAGTLKK823 pKa = 10.64LYY825 pKa = 10.96AGDD828 pKa = 3.64TGIEE832 pKa = 3.83RR833 pKa = 11.84VVIGTGTDD841 pKa = 3.05AVANSEE847 pKa = 4.17GTVALNVDD855 pKa = 4.4ASPLSNAITMIGNAGANILTGTAFADD881 pKa = 3.7QMDD884 pKa = 4.52GGSGNDD890 pKa = 4.11LLNGQAGIDD899 pKa = 3.77LMDD902 pKa = 4.36GEE904 pKa = 4.99EE905 pKa = 4.51GSDD908 pKa = 2.93IYY910 pKa = 11.28LIGLAAAHH918 pKa = 6.0TAAEE922 pKa = 4.24IADD925 pKa = 4.01TGSSGVDD932 pKa = 3.19EE933 pKa = 4.6VRR935 pKa = 11.84FAATTASTLTLFAGDD950 pKa = 3.53TGIEE954 pKa = 3.89RR955 pKa = 11.84VVLGTGTGAVATSTGSAALNADD977 pKa = 3.18ASALVNAITLVGNAGANILTGTEE1000 pKa = 3.59FADD1003 pKa = 4.89RR1004 pKa = 11.84IEE1006 pKa = 4.72GGGGADD1012 pKa = 3.3QLFGGSGNDD1021 pKa = 3.48TLFGGLGNDD1030 pKa = 3.48TLTGGEE1036 pKa = 4.41GEE1038 pKa = 4.39DD1039 pKa = 3.34HH1040 pKa = 6.82FVLNTAANAANNRR1053 pKa = 11.84DD1054 pKa = 3.51VLVDD1058 pKa = 4.1FEE1060 pKa = 4.93SGADD1064 pKa = 3.73HH1065 pKa = 7.33IEE1067 pKa = 3.98LSLLVFKK1074 pKa = 11.11GLGKK1078 pKa = 7.36TTGVLGEE1085 pKa = 4.06AQFWAAADD1093 pKa = 3.61AVKK1096 pKa = 10.64GHH1098 pKa = 7.01DD1099 pKa = 3.36ADD1101 pKa = 5.4DD1102 pKa = 3.46RR1103 pKa = 11.84VIYY1106 pKa = 8.32NTSTGSLYY1114 pKa = 10.94YY1115 pKa = 10.66DD1116 pKa = 3.89ADD1118 pKa = 3.97GSGKK1122 pKa = 10.06GAAVEE1127 pKa = 4.0IAVLGLSTHH1136 pKa = 6.56PTLEE1140 pKa = 4.51HH1141 pKa = 7.09GDD1143 pKa = 3.72FLLII1147 pKa = 4.64
Molecular weight: 116.3 kDa
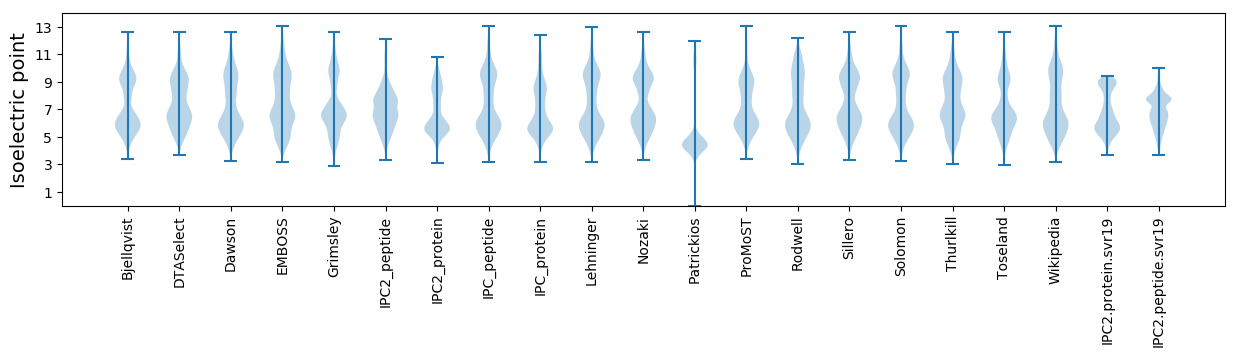
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A080MKE2|A0A080MKE2_9PROT Nitrite reductase [NAD(P)H] OS=Candidatus Accumulibacter sp. SK-02 OX=1453999 GN=nasD PE=4 SV=1
MM1 pKa = 7.61IGVLTSTPLAVALTVTLAGRR21 pKa = 11.84VWTVPSAGAVRR32 pKa = 11.84LTVGGVLPPFTVIVRR47 pKa = 11.84AALVVTPPALSVARR61 pKa = 11.84AVRR64 pKa = 11.84GWLPAGTLFQVKK76 pKa = 7.2VV77 pKa = 3.37
MM1 pKa = 7.61IGVLTSTPLAVALTVTLAGRR21 pKa = 11.84VWTVPSAGAVRR32 pKa = 11.84LTVGGVLPPFTVIVRR47 pKa = 11.84AALVVTPPALSVARR61 pKa = 11.84AVRR64 pKa = 11.84GWLPAGTLFQVKK76 pKa = 7.2VV77 pKa = 3.37
Molecular weight: 7.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291537 |
29 |
3115 |
326.7 |
35.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.845 ± 0.052 | 1.109 ± 0.014 |
5.323 ± 0.032 | 5.705 ± 0.041 |
3.641 ± 0.024 | 7.904 ± 0.045 |
2.304 ± 0.021 | 4.922 ± 0.03 |
3.076 ± 0.03 | 11.356 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.222 ± 0.018 | 2.688 ± 0.025 |
5.082 ± 0.03 | 3.894 ± 0.026 |
7.46 ± 0.046 | 5.645 ± 0.034 |
4.923 ± 0.039 | 7.165 ± 0.034 |
1.418 ± 0.017 | 2.315 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
