
Xanthomonas phage Xf109
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Xylivirus; Xanthomonas virus Xf109
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
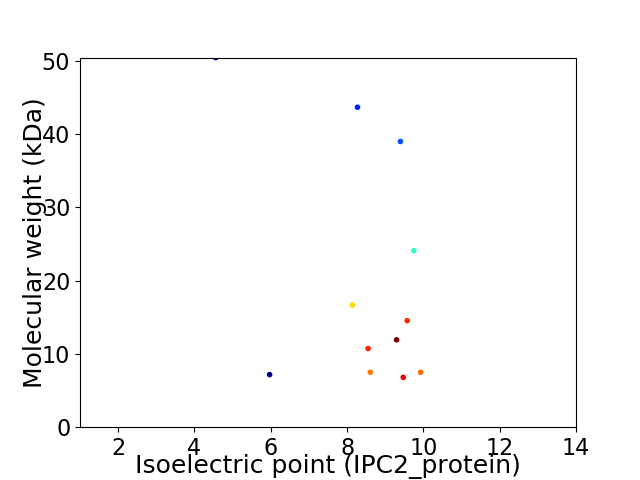
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1D6ZIU6|A0A1D6ZIU6_9VIRU DNA-Binding protein G5P OS=Xanthomonas phage Xf109 OX=1852597 GN=Xf109_2 PE=3 SV=1
MM1 pKa = 7.77AGGLRR6 pKa = 11.84DD7 pKa = 3.72CLRR10 pKa = 11.84GFRR13 pKa = 11.84IMGWLARR20 pKa = 11.84VFASAIARR28 pKa = 11.84RR29 pKa = 11.84LAYY32 pKa = 10.38VLVAATLAWCGIGCASAQDD51 pKa = 4.37FGTQGAAYY59 pKa = 8.25SACMSQTAAYY69 pKa = 8.79LASLGRR75 pKa = 11.84GANDD79 pKa = 3.87RR80 pKa = 11.84NPEE83 pKa = 4.17CNVEE87 pKa = 4.15QGSQAYY93 pKa = 9.58RR94 pKa = 11.84GQFEE98 pKa = 4.21TRR100 pKa = 11.84DD101 pKa = 3.73CSSCDD106 pKa = 2.74WYY108 pKa = 10.62RR109 pKa = 11.84AYY111 pKa = 10.81YY112 pKa = 10.46GYY114 pKa = 10.51FPWSKK119 pKa = 11.11GCDD122 pKa = 3.37QEE124 pKa = 4.35PDD126 pKa = 3.69YY127 pKa = 11.04TGSGPWGTYY136 pKa = 9.79VGTARR141 pKa = 11.84NGSIGCRR148 pKa = 11.84NGCDD152 pKa = 3.55GVWFGNGDD160 pKa = 4.33DD161 pKa = 3.78TMTWNATGAICPKK174 pKa = 10.5DD175 pKa = 3.59PEE177 pKa = 4.47KK178 pKa = 10.67TCDD181 pKa = 3.58AMSGGYY187 pKa = 8.8GWNGYY192 pKa = 10.1LGVCEE197 pKa = 4.44PPPTEE202 pKa = 4.26EE203 pKa = 4.33CPEE206 pKa = 4.09GQVPDD211 pKa = 4.11GKK213 pKa = 10.9GGCSSNKK220 pKa = 9.59CPEE223 pKa = 3.96GMLLQADD230 pKa = 4.98GTCAPKK236 pKa = 10.83KK237 pKa = 9.58NDD239 pKa = 3.67CPAGQIRR246 pKa = 11.84SPSGSCLPGDD256 pKa = 4.19GQCAAGEE263 pKa = 4.2VRR265 pKa = 11.84GPDD268 pKa = 3.62GTCKK272 pKa = 10.24KK273 pKa = 10.69DD274 pKa = 3.18GDD276 pKa = 4.3GDD278 pKa = 4.23GEE280 pKa = 4.16PDD282 pKa = 3.55EE283 pKa = 5.35PGEE286 pKa = 4.14GDD288 pKa = 2.97KK289 pKa = 11.39SQFSGGDD296 pKa = 3.37SCDD299 pKa = 3.89SPPSCSGDD307 pKa = 3.87AIMCGQARR315 pKa = 11.84IQWRR319 pKa = 11.84IDD321 pKa = 3.14CNTRR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.65VNITGGSCASMPVCVGEE344 pKa = 3.95NCKK347 pKa = 10.32ALEE350 pKa = 3.9YY351 pKa = 10.77SQLLMQWRR359 pKa = 11.84AACALEE365 pKa = 4.09KK366 pKa = 10.52AANNSGGGTGNNADD380 pKa = 3.25VKK382 pKa = 10.58AIRR385 pKa = 11.84DD386 pKa = 4.11AITGNGTADD395 pKa = 2.97IGADD399 pKa = 3.86GKK401 pKa = 10.07PADD404 pKa = 4.27AFSDD408 pKa = 3.4EE409 pKa = 4.41SGYY412 pKa = 11.23GEE414 pKa = 4.09HH415 pKa = 7.42GYY417 pKa = 8.04PTGEE421 pKa = 4.5LDD423 pKa = 3.36THH425 pKa = 6.16GFGYY429 pKa = 10.55SRR431 pKa = 11.84TCPTIPDD438 pKa = 3.49VAVFGQTLHH447 pKa = 6.82FDD449 pKa = 3.53TSKK452 pKa = 9.79FCQWMVLGGQIVLVMASLVSLRR474 pKa = 11.84LMSQGGSAA482 pKa = 3.86
MM1 pKa = 7.77AGGLRR6 pKa = 11.84DD7 pKa = 3.72CLRR10 pKa = 11.84GFRR13 pKa = 11.84IMGWLARR20 pKa = 11.84VFASAIARR28 pKa = 11.84RR29 pKa = 11.84LAYY32 pKa = 10.38VLVAATLAWCGIGCASAQDD51 pKa = 4.37FGTQGAAYY59 pKa = 8.25SACMSQTAAYY69 pKa = 8.79LASLGRR75 pKa = 11.84GANDD79 pKa = 3.87RR80 pKa = 11.84NPEE83 pKa = 4.17CNVEE87 pKa = 4.15QGSQAYY93 pKa = 9.58RR94 pKa = 11.84GQFEE98 pKa = 4.21TRR100 pKa = 11.84DD101 pKa = 3.73CSSCDD106 pKa = 2.74WYY108 pKa = 10.62RR109 pKa = 11.84AYY111 pKa = 10.81YY112 pKa = 10.46GYY114 pKa = 10.51FPWSKK119 pKa = 11.11GCDD122 pKa = 3.37QEE124 pKa = 4.35PDD126 pKa = 3.69YY127 pKa = 11.04TGSGPWGTYY136 pKa = 9.79VGTARR141 pKa = 11.84NGSIGCRR148 pKa = 11.84NGCDD152 pKa = 3.55GVWFGNGDD160 pKa = 4.33DD161 pKa = 3.78TMTWNATGAICPKK174 pKa = 10.5DD175 pKa = 3.59PEE177 pKa = 4.47KK178 pKa = 10.67TCDD181 pKa = 3.58AMSGGYY187 pKa = 8.8GWNGYY192 pKa = 10.1LGVCEE197 pKa = 4.44PPPTEE202 pKa = 4.26EE203 pKa = 4.33CPEE206 pKa = 4.09GQVPDD211 pKa = 4.11GKK213 pKa = 10.9GGCSSNKK220 pKa = 9.59CPEE223 pKa = 3.96GMLLQADD230 pKa = 4.98GTCAPKK236 pKa = 10.83KK237 pKa = 9.58NDD239 pKa = 3.67CPAGQIRR246 pKa = 11.84SPSGSCLPGDD256 pKa = 4.19GQCAAGEE263 pKa = 4.2VRR265 pKa = 11.84GPDD268 pKa = 3.62GTCKK272 pKa = 10.24KK273 pKa = 10.69DD274 pKa = 3.18GDD276 pKa = 4.3GDD278 pKa = 4.23GEE280 pKa = 4.16PDD282 pKa = 3.55EE283 pKa = 5.35PGEE286 pKa = 4.14GDD288 pKa = 2.97KK289 pKa = 11.39SQFSGGDD296 pKa = 3.37SCDD299 pKa = 3.89SPPSCSGDD307 pKa = 3.87AIMCGQARR315 pKa = 11.84IQWRR319 pKa = 11.84IDD321 pKa = 3.14CNTRR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.65VNITGGSCASMPVCVGEE344 pKa = 3.95NCKK347 pKa = 10.32ALEE350 pKa = 3.9YY351 pKa = 10.77SQLLMQWRR359 pKa = 11.84AACALEE365 pKa = 4.09KK366 pKa = 10.52AANNSGGGTGNNADD380 pKa = 3.25VKK382 pKa = 10.58AIRR385 pKa = 11.84DD386 pKa = 4.11AITGNGTADD395 pKa = 2.97IGADD399 pKa = 3.86GKK401 pKa = 10.07PADD404 pKa = 4.27AFSDD408 pKa = 3.4EE409 pKa = 4.41SGYY412 pKa = 11.23GEE414 pKa = 4.09HH415 pKa = 7.42GYY417 pKa = 8.04PTGEE421 pKa = 4.5LDD423 pKa = 3.36THH425 pKa = 6.16GFGYY429 pKa = 10.55SRR431 pKa = 11.84TCPTIPDD438 pKa = 3.49VAVFGQTLHH447 pKa = 6.82FDD449 pKa = 3.53TSKK452 pKa = 9.79FCQWMVLGGQIVLVMASLVSLRR474 pKa = 11.84LMSQGGSAA482 pKa = 3.86
Molecular weight: 50.4 kDa
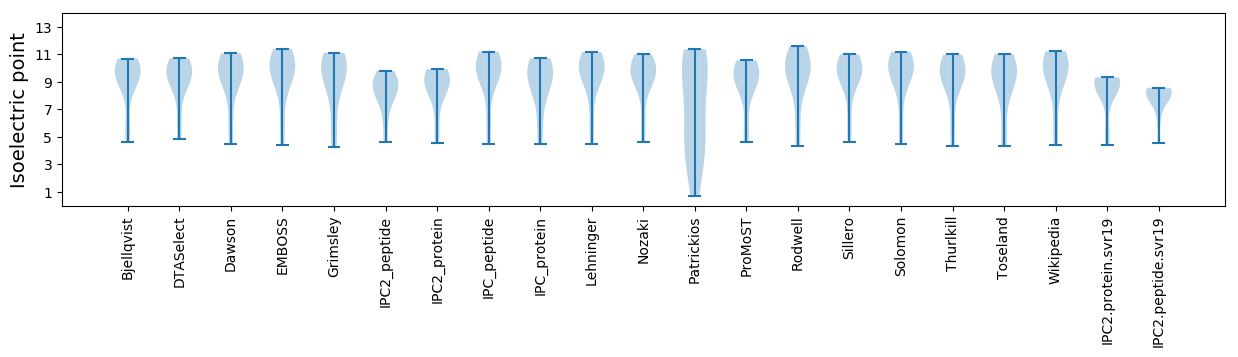
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D6ZIV1|A0A1D6ZIV1_9VIRU Uncharacterized protein OS=Xanthomonas phage Xf109 OX=1852597 GN=Xf109_11 PE=4 SV=1
MM1 pKa = 7.43TFDD4 pKa = 2.89TYY6 pKa = 11.55EE7 pKa = 4.21RR8 pKa = 11.84VDD10 pKa = 4.05LTGPWAGFGFQGHH23 pKa = 6.07RR24 pKa = 11.84FFTPEE29 pKa = 4.2NYY31 pKa = 10.47DD32 pKa = 3.09IDD34 pKa = 4.44PCGMRR39 pKa = 11.84YY40 pKa = 8.0WALTCAIARR49 pKa = 11.84EE50 pKa = 4.08WSLMMSEE57 pKa = 4.29EE58 pKa = 4.61RR59 pKa = 11.84NAQSANPRR67 pKa = 11.84TPTATRR73 pKa = 11.84SPGSRR78 pKa = 11.84ISEE81 pKa = 4.28DD82 pKa = 3.26ANVIYY87 pKa = 10.62LRR89 pKa = 11.84DD90 pKa = 3.66VLWRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84EE97 pKa = 3.78KK98 pKa = 10.64RR99 pKa = 11.84LSVGGNPGLADD110 pKa = 3.44SGKK113 pKa = 10.14RR114 pKa = 11.84SPTGRR119 pKa = 11.84GRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84PRR125 pKa = 11.84RR126 pKa = 11.84GG127 pKa = 2.87
MM1 pKa = 7.43TFDD4 pKa = 2.89TYY6 pKa = 11.55EE7 pKa = 4.21RR8 pKa = 11.84VDD10 pKa = 4.05LTGPWAGFGFQGHH23 pKa = 6.07RR24 pKa = 11.84FFTPEE29 pKa = 4.2NYY31 pKa = 10.47DD32 pKa = 3.09IDD34 pKa = 4.44PCGMRR39 pKa = 11.84YY40 pKa = 8.0WALTCAIARR49 pKa = 11.84EE50 pKa = 4.08WSLMMSEE57 pKa = 4.29EE58 pKa = 4.61RR59 pKa = 11.84NAQSANPRR67 pKa = 11.84TPTATRR73 pKa = 11.84SPGSRR78 pKa = 11.84ISEE81 pKa = 4.28DD82 pKa = 3.26ANVIYY87 pKa = 10.62LRR89 pKa = 11.84DD90 pKa = 3.66VLWRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84EE97 pKa = 3.78KK98 pKa = 10.64RR99 pKa = 11.84LSVGGNPGLADD110 pKa = 3.44SGKK113 pKa = 10.14RR114 pKa = 11.84SPTGRR119 pKa = 11.84GRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84PRR125 pKa = 11.84RR126 pKa = 11.84GG127 pKa = 2.87
Molecular weight: 14.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2202 |
61 |
482 |
183.5 |
19.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.398 ± 0.882 | 2.316 ± 0.929 |
5.086 ± 0.715 | 3.724 ± 0.492 |
3.815 ± 0.654 | 9.991 ± 1.204 |
1.635 ± 0.325 | 3.951 ± 0.5 |
4.042 ± 0.653 | 8.447 ± 1.513 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.543 ± 0.279 | 2.316 ± 0.339 |
4.905 ± 0.603 | 3.951 ± 0.623 |
7.039 ± 0.947 | 5.631 ± 0.551 |
5.858 ± 0.756 | 6.63 ± 1.124 |
2.861 ± 0.494 | 2.861 ± 0.324 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
