
Phaeomoniella chlamydospora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Phaeomoniellales; Phaeomoniellaceae; Phaeomoniella
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
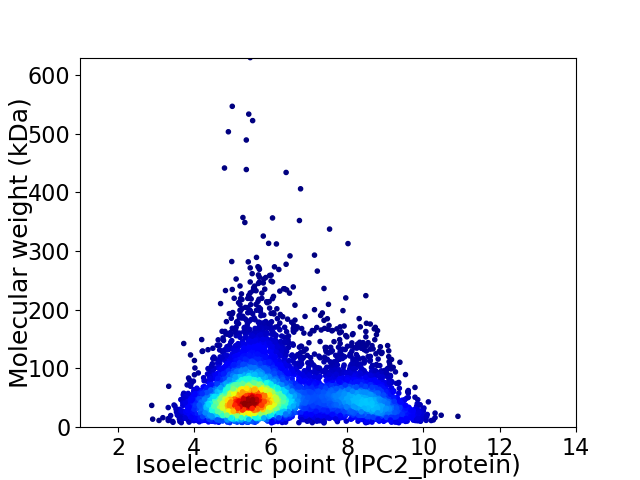
Virtual 2D-PAGE plot for 6907 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2E4V8|A0A0G2E4V8_9EURO Putative fes cip4 homology domain protein OS=Phaeomoniella chlamydospora OX=158046 GN=UCRPC4_g05387 PE=4 SV=1
MM1 pKa = 7.77PSYY4 pKa = 10.58KK5 pKa = 10.25FSLAVIALSSASAVLSNPIAAPAASPAPTSAPDD38 pKa = 3.18INKK41 pKa = 9.83RR42 pKa = 11.84ATTCTFTGSKK52 pKa = 9.84GASFASKK59 pKa = 10.32SKK61 pKa = 10.38SSCSTIVLSAVAVPSGTTLDD81 pKa = 3.84LTDD84 pKa = 5.38LEE86 pKa = 5.69DD87 pKa = 3.97GTHH90 pKa = 6.7TDD92 pKa = 5.8LIMTDD97 pKa = 3.07ITIDD101 pKa = 4.02NSDD104 pKa = 3.6GDD106 pKa = 4.54DD107 pKa = 3.42NGAANTDD114 pKa = 3.35GFDD117 pKa = 3.51IGDD120 pKa = 3.73STGFTIKK127 pKa = 10.53SATVYY132 pKa = 10.12NQHH135 pKa = 6.91DD136 pKa = 4.0CVAINSGEE144 pKa = 4.18TIVFSGGYY152 pKa = 9.47CSGGHH157 pKa = 5.68GLSVGSVGGRR167 pKa = 11.84SDD169 pKa = 3.5NTVKK173 pKa = 10.7GVTFEE178 pKa = 4.06SSTVINSQNGVRR190 pKa = 11.84IKK192 pKa = 9.73TVYY195 pKa = 10.39DD196 pKa = 3.24ATGEE200 pKa = 4.39VTDD203 pKa = 3.97VTYY206 pKa = 11.33KK207 pKa = 10.93DD208 pKa = 3.26ITLSGITKK216 pKa = 8.48YY217 pKa = 10.97GIVIRR222 pKa = 11.84QDD224 pKa = 3.55YY225 pKa = 10.69EE226 pKa = 4.08NGSPTGDD233 pKa = 3.57PTTGVPITDD242 pKa = 3.71CTFDD246 pKa = 4.26EE247 pKa = 4.27ITGTVEE253 pKa = 3.51SDD255 pKa = 2.95AVEE258 pKa = 4.08IFILCGDD265 pKa = 4.83DD266 pKa = 4.87SCSDD270 pKa = 3.61WTWEE274 pKa = 5.14DD275 pKa = 2.81IDD277 pKa = 3.69ITGGEE282 pKa = 4.21TSDD285 pKa = 3.46GCEE288 pKa = 3.96NVPSGASCSEE298 pKa = 3.98
MM1 pKa = 7.77PSYY4 pKa = 10.58KK5 pKa = 10.25FSLAVIALSSASAVLSNPIAAPAASPAPTSAPDD38 pKa = 3.18INKK41 pKa = 9.83RR42 pKa = 11.84ATTCTFTGSKK52 pKa = 9.84GASFASKK59 pKa = 10.32SKK61 pKa = 10.38SSCSTIVLSAVAVPSGTTLDD81 pKa = 3.84LTDD84 pKa = 5.38LEE86 pKa = 5.69DD87 pKa = 3.97GTHH90 pKa = 6.7TDD92 pKa = 5.8LIMTDD97 pKa = 3.07ITIDD101 pKa = 4.02NSDD104 pKa = 3.6GDD106 pKa = 4.54DD107 pKa = 3.42NGAANTDD114 pKa = 3.35GFDD117 pKa = 3.51IGDD120 pKa = 3.73STGFTIKK127 pKa = 10.53SATVYY132 pKa = 10.12NQHH135 pKa = 6.91DD136 pKa = 4.0CVAINSGEE144 pKa = 4.18TIVFSGGYY152 pKa = 9.47CSGGHH157 pKa = 5.68GLSVGSVGGRR167 pKa = 11.84SDD169 pKa = 3.5NTVKK173 pKa = 10.7GVTFEE178 pKa = 4.06SSTVINSQNGVRR190 pKa = 11.84IKK192 pKa = 9.73TVYY195 pKa = 10.39DD196 pKa = 3.24ATGEE200 pKa = 4.39VTDD203 pKa = 3.97VTYY206 pKa = 11.33KK207 pKa = 10.93DD208 pKa = 3.26ITLSGITKK216 pKa = 8.48YY217 pKa = 10.97GIVIRR222 pKa = 11.84QDD224 pKa = 3.55YY225 pKa = 10.69EE226 pKa = 4.08NGSPTGDD233 pKa = 3.57PTTGVPITDD242 pKa = 3.71CTFDD246 pKa = 4.26EE247 pKa = 4.27ITGTVEE253 pKa = 3.51SDD255 pKa = 2.95AVEE258 pKa = 4.08IFILCGDD265 pKa = 4.83DD266 pKa = 4.87SCSDD270 pKa = 3.61WTWEE274 pKa = 5.14DD275 pKa = 2.81IDD277 pKa = 3.69ITGGEE282 pKa = 4.21TSDD285 pKa = 3.46GCEE288 pKa = 3.96NVPSGASCSEE298 pKa = 3.98
Molecular weight: 30.54 kDa
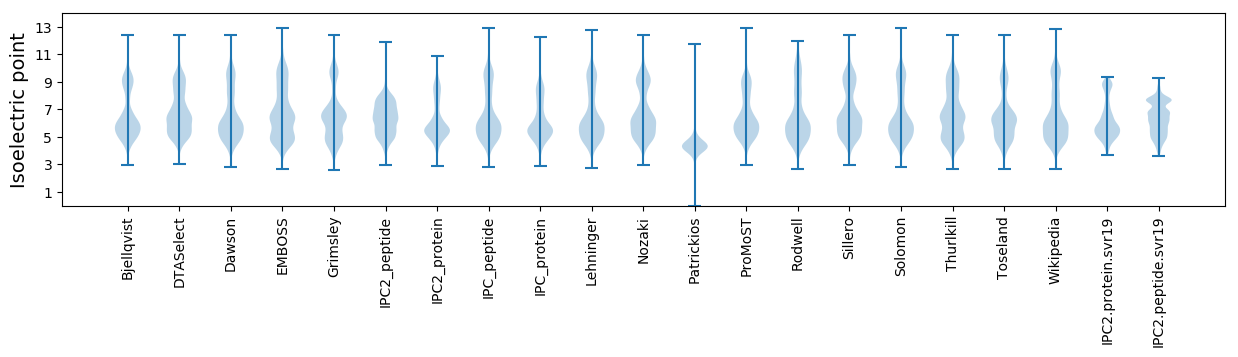
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2F1E5|A0A0G2F1E5_9EURO Putative exportin OS=Phaeomoniella chlamydospora OX=158046 GN=UCRPC4_g00438 PE=3 SV=1
MM1 pKa = 7.69ASTRR5 pKa = 11.84SLYY8 pKa = 11.08KK9 pKa = 9.68NTADD13 pKa = 4.08DD14 pKa = 3.69RR15 pKa = 11.84SGIIIVITYY24 pKa = 10.5VMLSVSTFLLLLQAGVGIYY43 pKa = 10.22LKK45 pKa = 10.53HH46 pKa = 6.21PKK48 pKa = 9.77GVNDD52 pKa = 4.04ILLHH56 pKa = 5.45LAGVFSIAQSIAITLSATNGLGRR79 pKa = 11.84HH80 pKa = 5.14RR81 pKa = 11.84HH82 pKa = 5.87DD83 pKa = 4.65LSNGQFNRR91 pKa = 11.84FSKK94 pKa = 10.54FYY96 pKa = 10.29YY97 pKa = 10.13AGQICQIVAVGLSKK111 pKa = 10.97LSVIIIINRR120 pKa = 11.84LSSNRR125 pKa = 11.84KK126 pKa = 8.72VILACRR132 pKa = 11.84CFAVVIIIYY141 pKa = 10.19ILFSTLTTAFQCRR154 pKa = 11.84FPFWTFAPNRR164 pKa = 11.84CVLRR168 pKa = 11.84GNLLYY173 pKa = 10.76AISDD177 pKa = 3.95SNALLDD183 pKa = 3.51IVLIILPIVAIIQYY197 pKa = 10.13KK198 pKa = 9.59PRR200 pKa = 11.84LPQRR204 pKa = 11.84GFLIASFITRR214 pKa = 11.84AAPLAAAILSLVSLPTYY231 pKa = 10.28LHH233 pKa = 5.47NTKK236 pKa = 9.3DD237 pKa = 3.92TTWTILPISTINEE250 pKa = 3.92ILVHH254 pKa = 6.01LNIIVACIPSILLISLNPPLPSNRR278 pKa = 11.84EE279 pKa = 3.84TASNNPIHH287 pKa = 6.98RR288 pKa = 11.84HH289 pKa = 5.32RR290 pKa = 11.84PPPFQKK296 pKa = 10.62LPSHH300 pKa = 6.7TNTTIRR306 pKa = 11.84PSTPLFRR313 pKa = 11.84YY314 pKa = 8.53AHH316 pKa = 6.26NFSPIQSFFHH326 pKa = 6.41ISGGRR331 pKa = 11.84EE332 pKa = 3.64ACDD335 pKa = 3.2NNNNRR340 pKa = 11.84RR341 pKa = 11.84SDD343 pKa = 3.34RR344 pKa = 11.84RR345 pKa = 11.84GPNNNNNTSKK355 pKa = 9.17YY356 pKa = 7.84TGGGGVAGRR365 pKa = 11.84RR366 pKa = 11.84RR367 pKa = 11.84RR368 pKa = 11.84GDD370 pKa = 3.1EE371 pKa = 4.03DD372 pKa = 3.79NNNNTTRR379 pKa = 11.84SPTKK383 pKa = 9.31RR384 pKa = 11.84KK385 pKa = 10.51SMADD389 pKa = 3.25SLRR392 pKa = 11.84TLTSFKK398 pKa = 10.59GVRR401 pKa = 11.84KK402 pKa = 7.56TMEE405 pKa = 3.54VRR407 pKa = 11.84VEE409 pKa = 3.63IEE411 pKa = 3.6EE412 pKa = 4.31RR413 pKa = 11.84GVSRR417 pKa = 11.84RR418 pKa = 11.84GGGIGGG424 pKa = 3.68
MM1 pKa = 7.69ASTRR5 pKa = 11.84SLYY8 pKa = 11.08KK9 pKa = 9.68NTADD13 pKa = 4.08DD14 pKa = 3.69RR15 pKa = 11.84SGIIIVITYY24 pKa = 10.5VMLSVSTFLLLLQAGVGIYY43 pKa = 10.22LKK45 pKa = 10.53HH46 pKa = 6.21PKK48 pKa = 9.77GVNDD52 pKa = 4.04ILLHH56 pKa = 5.45LAGVFSIAQSIAITLSATNGLGRR79 pKa = 11.84HH80 pKa = 5.14RR81 pKa = 11.84HH82 pKa = 5.87DD83 pKa = 4.65LSNGQFNRR91 pKa = 11.84FSKK94 pKa = 10.54FYY96 pKa = 10.29YY97 pKa = 10.13AGQICQIVAVGLSKK111 pKa = 10.97LSVIIIINRR120 pKa = 11.84LSSNRR125 pKa = 11.84KK126 pKa = 8.72VILACRR132 pKa = 11.84CFAVVIIIYY141 pKa = 10.19ILFSTLTTAFQCRR154 pKa = 11.84FPFWTFAPNRR164 pKa = 11.84CVLRR168 pKa = 11.84GNLLYY173 pKa = 10.76AISDD177 pKa = 3.95SNALLDD183 pKa = 3.51IVLIILPIVAIIQYY197 pKa = 10.13KK198 pKa = 9.59PRR200 pKa = 11.84LPQRR204 pKa = 11.84GFLIASFITRR214 pKa = 11.84AAPLAAAILSLVSLPTYY231 pKa = 10.28LHH233 pKa = 5.47NTKK236 pKa = 9.3DD237 pKa = 3.92TTWTILPISTINEE250 pKa = 3.92ILVHH254 pKa = 6.01LNIIVACIPSILLISLNPPLPSNRR278 pKa = 11.84EE279 pKa = 3.84TASNNPIHH287 pKa = 6.98RR288 pKa = 11.84HH289 pKa = 5.32RR290 pKa = 11.84PPPFQKK296 pKa = 10.62LPSHH300 pKa = 6.7TNTTIRR306 pKa = 11.84PSTPLFRR313 pKa = 11.84YY314 pKa = 8.53AHH316 pKa = 6.26NFSPIQSFFHH326 pKa = 6.41ISGGRR331 pKa = 11.84EE332 pKa = 3.64ACDD335 pKa = 3.2NNNNRR340 pKa = 11.84RR341 pKa = 11.84SDD343 pKa = 3.34RR344 pKa = 11.84RR345 pKa = 11.84GPNNNNNTSKK355 pKa = 9.17YY356 pKa = 7.84TGGGGVAGRR365 pKa = 11.84RR366 pKa = 11.84RR367 pKa = 11.84RR368 pKa = 11.84GDD370 pKa = 3.1EE371 pKa = 4.03DD372 pKa = 3.79NNNNTTRR379 pKa = 11.84SPTKK383 pKa = 9.31RR384 pKa = 11.84KK385 pKa = 10.51SMADD389 pKa = 3.25SLRR392 pKa = 11.84TLTSFKK398 pKa = 10.59GVRR401 pKa = 11.84KK402 pKa = 7.56TMEE405 pKa = 3.54VRR407 pKa = 11.84VEE409 pKa = 3.63IEE411 pKa = 3.6EE412 pKa = 4.31RR413 pKa = 11.84GVSRR417 pKa = 11.84RR418 pKa = 11.84GGGIGGG424 pKa = 3.68
Molecular weight: 46.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3771177 |
66 |
5663 |
546.0 |
60.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.238 ± 0.021 | 1.019 ± 0.008 |
5.933 ± 0.018 | 6.469 ± 0.025 |
3.711 ± 0.017 | 6.816 ± 0.027 |
2.272 ± 0.012 | 5.284 ± 0.019 |
5.281 ± 0.022 | 8.801 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.082 ± 0.01 | 3.888 ± 0.015 |
5.699 ± 0.027 | 4.031 ± 0.018 |
5.742 ± 0.025 | 8.427 ± 0.03 |
6.182 ± 0.022 | 5.973 ± 0.018 |
1.346 ± 0.009 | 2.804 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
