
Trametes versicolor (strain FP-101664) (White-rot fungus) (Coriolus versicolor)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Trametes; Trametes versicolor
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
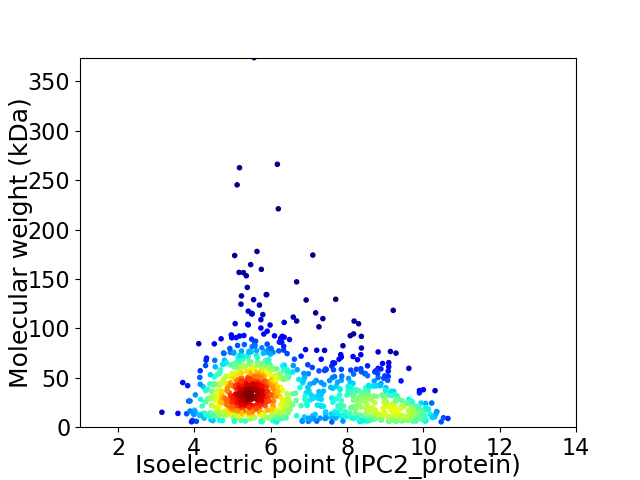
Virtual 2D-PAGE plot for 1022 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
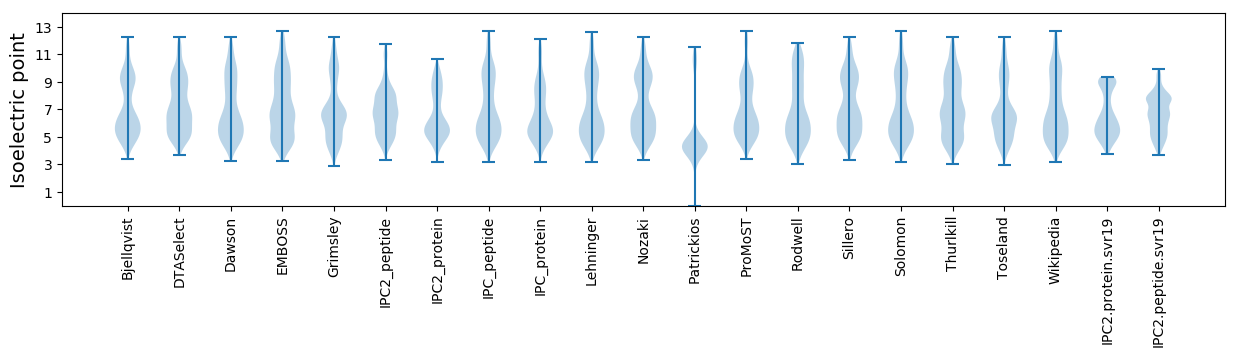
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7S7K5|R7S7K5_TRAVS Phosphoglucomutase OS=Trametes versicolor (strain FP-101664) OX=717944 GN=TRAVEDRAFT_157183 PE=3 SV=1
LL1 pKa = 7.11LATIISLFFASAPVGAFAVLQIDD24 pKa = 4.33TPSPAPTQCAPAILQWSGGAPPYY47 pKa = 10.34FLAIVGTGDD56 pKa = 4.19LAVALQRR63 pKa = 11.84YY64 pKa = 9.23DD65 pKa = 3.75GLFDD69 pKa = 4.42TSFLWSTNITARR81 pKa = 11.84TSIALTVTDD90 pKa = 3.5VTGTLAQSAPFIIHH104 pKa = 6.14SSSDD108 pKa = 3.25ISCLEE113 pKa = 3.78SSVPSSSDD121 pKa = 2.41ISRR124 pKa = 11.84VLDD127 pKa = 3.35STTFEE132 pKa = 4.81ASTSMFPAGITAGASAGVSGGAGATAVATAGVV164 pKa = 3.56
LL1 pKa = 7.11LATIISLFFASAPVGAFAVLQIDD24 pKa = 4.33TPSPAPTQCAPAILQWSGGAPPYY47 pKa = 10.34FLAIVGTGDD56 pKa = 4.19LAVALQRR63 pKa = 11.84YY64 pKa = 9.23DD65 pKa = 3.75GLFDD69 pKa = 4.42TSFLWSTNITARR81 pKa = 11.84TSIALTVTDD90 pKa = 3.5VTGTLAQSAPFIIHH104 pKa = 6.14SSSDD108 pKa = 3.25ISCLEE113 pKa = 3.78SSVPSSSDD121 pKa = 2.41ISRR124 pKa = 11.84VLDD127 pKa = 3.35STTFEE132 pKa = 4.81ASTSMFPAGITAGASAGVSGGAGATAVATAGVV164 pKa = 3.56
Molecular weight: 16.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7S788|R7S788_TRAVS Nitric oxide dioxygenase OS=Trametes versicolor (strain FP-101664) OX=717944 GN=TRAVEDRAFT_136661 PE=3 SV=1
MM1 pKa = 7.8PSLRR5 pKa = 11.84LPFSCMALSLLDD17 pKa = 3.47GHH19 pKa = 6.85TDD21 pKa = 3.31TSSPAALAAHH31 pKa = 6.05TRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PSPASSTLPEE45 pKa = 4.05SLAPYY50 pKa = 7.53STPLDD55 pKa = 4.16DD56 pKa = 3.89PTTPRR61 pKa = 11.84TSLRR65 pKa = 11.84LWDD68 pKa = 3.73LHH70 pKa = 6.37SALSFALLDD79 pKa = 3.89VLDD82 pKa = 4.52VPAAPSLGQQAGLASRR98 pKa = 11.84KK99 pKa = 5.27TTPFGAANLARR110 pKa = 11.84IMHH113 pKa = 7.15RR114 pKa = 11.84STPRR118 pKa = 11.84PQRR121 pKa = 11.84AASGKK126 pKa = 8.05NWQVLASYY134 pKa = 7.95STSWDD139 pKa = 3.07RR140 pKa = 11.84LRR142 pKa = 11.84ALTSTGSRR150 pKa = 11.84KK151 pKa = 9.33NASDD155 pKa = 3.95PRR157 pKa = 11.84SCSQTRR163 pKa = 11.84IWAHH167 pKa = 6.0HH168 pKa = 5.43GPNRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AICAANTDD182 pKa = 3.56SCDD185 pKa = 3.45ARR187 pKa = 11.84QHH189 pKa = 5.33SQHH192 pKa = 6.24TSSGIQPAYY201 pKa = 10.48RR202 pKa = 11.84PGSPPAPKK210 pKa = 9.03TANTHH215 pKa = 4.34HH216 pKa = 7.43TYY218 pKa = 8.71THH220 pKa = 6.17TCQTILHH227 pKa = 7.06ALQDD231 pKa = 3.51VALAPKK237 pKa = 10.11AVADD241 pKa = 3.98RR242 pKa = 11.84CFTILII248 pKa = 4.4
MM1 pKa = 7.8PSLRR5 pKa = 11.84LPFSCMALSLLDD17 pKa = 3.47GHH19 pKa = 6.85TDD21 pKa = 3.31TSSPAALAAHH31 pKa = 6.05TRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PSPASSTLPEE45 pKa = 4.05SLAPYY50 pKa = 7.53STPLDD55 pKa = 4.16DD56 pKa = 3.89PTTPRR61 pKa = 11.84TSLRR65 pKa = 11.84LWDD68 pKa = 3.73LHH70 pKa = 6.37SALSFALLDD79 pKa = 3.89VLDD82 pKa = 4.52VPAAPSLGQQAGLASRR98 pKa = 11.84KK99 pKa = 5.27TTPFGAANLARR110 pKa = 11.84IMHH113 pKa = 7.15RR114 pKa = 11.84STPRR118 pKa = 11.84PQRR121 pKa = 11.84AASGKK126 pKa = 8.05NWQVLASYY134 pKa = 7.95STSWDD139 pKa = 3.07RR140 pKa = 11.84LRR142 pKa = 11.84ALTSTGSRR150 pKa = 11.84KK151 pKa = 9.33NASDD155 pKa = 3.95PRR157 pKa = 11.84SCSQTRR163 pKa = 11.84IWAHH167 pKa = 6.0HH168 pKa = 5.43GPNRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AICAANTDD182 pKa = 3.56SCDD185 pKa = 3.45ARR187 pKa = 11.84QHH189 pKa = 5.33SQHH192 pKa = 6.24TSSGIQPAYY201 pKa = 10.48RR202 pKa = 11.84PGSPPAPKK210 pKa = 9.03TANTHH215 pKa = 4.34HH216 pKa = 7.43TYY218 pKa = 8.71THH220 pKa = 6.17TCQTILHH227 pKa = 7.06ALQDD231 pKa = 3.51VALAPKK237 pKa = 10.11AVADD241 pKa = 3.98RR242 pKa = 11.84CFTILII248 pKa = 4.4
Molecular weight: 26.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
359438 |
49 |
3380 |
351.7 |
38.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.894 ± 0.087 | 1.34 ± 0.034 |
5.96 ± 0.064 | 5.916 ± 0.08 |
3.52 ± 0.054 | 6.47 ± 0.075 |
2.742 ± 0.04 | 4.323 ± 0.054 |
3.904 ± 0.065 | 9.022 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.064 ± 0.033 | 2.923 ± 0.038 |
6.912 ± 0.1 | 3.593 ± 0.053 |
6.934 ± 0.069 | 7.803 ± 0.098 |
6.029 ± 0.061 | 6.49 ± 0.062 |
1.535 ± 0.031 | 2.628 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
