
Sediminispirochaeta smaragdinae (strain DSM 11293 / JCM 15392 / SEBR 4228) (Spirochaeta smaragdinae)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Sediminispirochaeta; Sediminispirochaeta smaragdinae
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
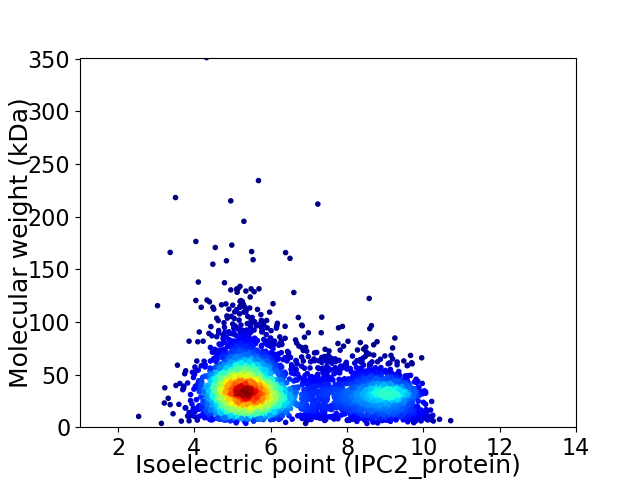
Virtual 2D-PAGE plot for 4211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1R7Y3|E1R7Y3_SEDSS 30S ribosomal protein S9 OS=Sediminispirochaeta smaragdinae (strain DSM 11293 / JCM 15392 / SEBR 4228) OX=573413 GN=rpsI PE=3 SV=1
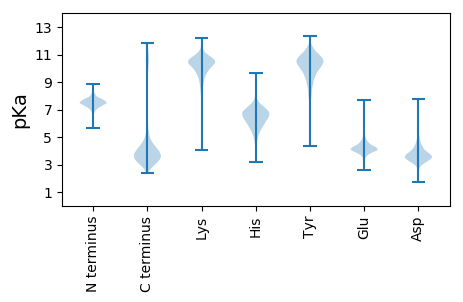
MM1 pKa = 7.5SSLRR5 pKa = 11.84RR6 pKa = 11.84ITTVAFTAAALLLAGVSLSADD27 pKa = 3.38DD28 pKa = 5.57FSFDD32 pKa = 5.03DD33 pKa = 4.48FGSSDD38 pKa = 3.61FGDD41 pKa = 3.79DD42 pKa = 3.27TGTATAGSSSAIEE55 pKa = 3.9VHH57 pKa = 6.6GEE59 pKa = 3.63LDD61 pKa = 3.18GSLRR65 pKa = 11.84ATVGEE70 pKa = 4.71DD71 pKa = 3.01ALSDD75 pKa = 3.08IGAISSEE82 pKa = 4.15EE83 pKa = 3.71DD84 pKa = 3.31SIEE87 pKa = 4.14YY88 pKa = 10.53GSEE91 pKa = 3.59AKK93 pKa = 10.8LNLTYY98 pKa = 10.65TGDD101 pKa = 3.81TIDD104 pKa = 5.14AFINLQIDD112 pKa = 3.75PEE114 pKa = 4.08RR115 pKa = 11.84LEE117 pKa = 6.12DD118 pKa = 5.05DD119 pKa = 3.2MSTLFDD125 pKa = 3.43EE126 pKa = 5.77AYY128 pKa = 9.22IRR130 pKa = 11.84YY131 pKa = 8.86YY132 pKa = 11.11GSGFDD137 pKa = 5.01FEE139 pKa = 4.66TGLMKK144 pKa = 10.27VVWGKK149 pKa = 10.95GDD151 pKa = 3.38QLHH154 pKa = 6.5VVDD157 pKa = 5.89LLNSDD162 pKa = 4.73DD163 pKa = 4.47LTDD166 pKa = 4.75FVNPDD171 pKa = 3.51YY172 pKa = 10.73IDD174 pKa = 3.52RR175 pKa = 11.84RR176 pKa = 11.84LAVPMAKK183 pKa = 10.3LNVQVGVQGKK193 pKa = 9.84LEE195 pKa = 4.06FVYY198 pKa = 10.58IPTLTPNTNPTSGAWATQQAQDD220 pKa = 4.3LVDD223 pKa = 3.42KK224 pKa = 9.73TEE226 pKa = 3.86EE227 pKa = 4.3YY228 pKa = 8.39ITYY231 pKa = 10.17VALSAYY237 pKa = 10.29NEE239 pKa = 4.61TYY241 pKa = 9.53TATSSEE247 pKa = 4.1AAATLAEE254 pKa = 3.96ASILEE259 pKa = 4.34KK260 pKa = 11.16YY261 pKa = 10.62SDD263 pKa = 3.86GSALYY268 pKa = 9.76PDD270 pKa = 5.17TDD272 pKa = 3.72TLDD275 pKa = 3.49YY276 pKa = 11.07SQAAVRR282 pKa = 11.84WTDD285 pKa = 2.99SFGGIDD291 pKa = 3.75VGALYY296 pKa = 11.07YY297 pKa = 10.77CGFLKK302 pKa = 10.71DD303 pKa = 3.75PSISYY308 pKa = 8.75TVDD311 pKa = 2.94GSGTPTAIDD320 pKa = 3.09ISYY323 pKa = 10.97DD324 pKa = 3.29RR325 pKa = 11.84LHH327 pKa = 7.1AFGLEE332 pKa = 3.69AGGIVGGFNLRR343 pKa = 11.84SEE345 pKa = 4.27AAYY348 pKa = 11.4YY349 pKa = 6.8MTEE352 pKa = 5.04DD353 pKa = 3.44IAGDD357 pKa = 3.75DD358 pKa = 3.59PVVHH362 pKa = 6.48NNSLNYY368 pKa = 10.43LVGFDD373 pKa = 4.92RR374 pKa = 11.84DD375 pKa = 3.86LPLHH379 pKa = 6.32NINLNMQATGSYY391 pKa = 10.03ILNNDD396 pKa = 3.94EE397 pKa = 4.61ISNGDD402 pKa = 3.3IEE404 pKa = 5.17YY405 pKa = 10.87DD406 pKa = 3.51GDD408 pKa = 4.48DD409 pKa = 4.74DD410 pKa = 4.67YY411 pKa = 12.15SSNLIVCKK419 pKa = 10.67ISDD422 pKa = 3.91TFNHH426 pKa = 5.92EE427 pKa = 4.2KK428 pKa = 10.77VSIEE432 pKa = 3.63AKK434 pKa = 10.15GIYY437 pKa = 9.66EE438 pKa = 4.69IEE440 pKa = 4.77DD441 pKa = 4.18EE442 pKa = 4.4DD443 pKa = 3.99WMIGPEE449 pKa = 3.83ITLAPNGEE457 pKa = 4.36FEE459 pKa = 4.55FSAEE463 pKa = 3.66ARR465 pKa = 11.84FFGGDD470 pKa = 3.16DD471 pKa = 4.1DD472 pKa = 5.2GRR474 pKa = 11.84LGQFDD479 pKa = 4.05EE480 pKa = 4.72SSYY483 pKa = 11.41LQFKK487 pKa = 8.9VKK489 pKa = 10.83AMFF492 pKa = 4.09
MM1 pKa = 7.5SSLRR5 pKa = 11.84RR6 pKa = 11.84ITTVAFTAAALLLAGVSLSADD27 pKa = 3.38DD28 pKa = 5.57FSFDD32 pKa = 5.03DD33 pKa = 4.48FGSSDD38 pKa = 3.61FGDD41 pKa = 3.79DD42 pKa = 3.27TGTATAGSSSAIEE55 pKa = 3.9VHH57 pKa = 6.6GEE59 pKa = 3.63LDD61 pKa = 3.18GSLRR65 pKa = 11.84ATVGEE70 pKa = 4.71DD71 pKa = 3.01ALSDD75 pKa = 3.08IGAISSEE82 pKa = 4.15EE83 pKa = 3.71DD84 pKa = 3.31SIEE87 pKa = 4.14YY88 pKa = 10.53GSEE91 pKa = 3.59AKK93 pKa = 10.8LNLTYY98 pKa = 10.65TGDD101 pKa = 3.81TIDD104 pKa = 5.14AFINLQIDD112 pKa = 3.75PEE114 pKa = 4.08RR115 pKa = 11.84LEE117 pKa = 6.12DD118 pKa = 5.05DD119 pKa = 3.2MSTLFDD125 pKa = 3.43EE126 pKa = 5.77AYY128 pKa = 9.22IRR130 pKa = 11.84YY131 pKa = 8.86YY132 pKa = 11.11GSGFDD137 pKa = 5.01FEE139 pKa = 4.66TGLMKK144 pKa = 10.27VVWGKK149 pKa = 10.95GDD151 pKa = 3.38QLHH154 pKa = 6.5VVDD157 pKa = 5.89LLNSDD162 pKa = 4.73DD163 pKa = 4.47LTDD166 pKa = 4.75FVNPDD171 pKa = 3.51YY172 pKa = 10.73IDD174 pKa = 3.52RR175 pKa = 11.84RR176 pKa = 11.84LAVPMAKK183 pKa = 10.3LNVQVGVQGKK193 pKa = 9.84LEE195 pKa = 4.06FVYY198 pKa = 10.58IPTLTPNTNPTSGAWATQQAQDD220 pKa = 4.3LVDD223 pKa = 3.42KK224 pKa = 9.73TEE226 pKa = 3.86EE227 pKa = 4.3YY228 pKa = 8.39ITYY231 pKa = 10.17VALSAYY237 pKa = 10.29NEE239 pKa = 4.61TYY241 pKa = 9.53TATSSEE247 pKa = 4.1AAATLAEE254 pKa = 3.96ASILEE259 pKa = 4.34KK260 pKa = 11.16YY261 pKa = 10.62SDD263 pKa = 3.86GSALYY268 pKa = 9.76PDD270 pKa = 5.17TDD272 pKa = 3.72TLDD275 pKa = 3.49YY276 pKa = 11.07SQAAVRR282 pKa = 11.84WTDD285 pKa = 2.99SFGGIDD291 pKa = 3.75VGALYY296 pKa = 11.07YY297 pKa = 10.77CGFLKK302 pKa = 10.71DD303 pKa = 3.75PSISYY308 pKa = 8.75TVDD311 pKa = 2.94GSGTPTAIDD320 pKa = 3.09ISYY323 pKa = 10.97DD324 pKa = 3.29RR325 pKa = 11.84LHH327 pKa = 7.1AFGLEE332 pKa = 3.69AGGIVGGFNLRR343 pKa = 11.84SEE345 pKa = 4.27AAYY348 pKa = 11.4YY349 pKa = 6.8MTEE352 pKa = 5.04DD353 pKa = 3.44IAGDD357 pKa = 3.75DD358 pKa = 3.59PVVHH362 pKa = 6.48NNSLNYY368 pKa = 10.43LVGFDD373 pKa = 4.92RR374 pKa = 11.84DD375 pKa = 3.86LPLHH379 pKa = 6.32NINLNMQATGSYY391 pKa = 10.03ILNNDD396 pKa = 3.94EE397 pKa = 4.61ISNGDD402 pKa = 3.3IEE404 pKa = 5.17YY405 pKa = 10.87DD406 pKa = 3.51GDD408 pKa = 4.48DD409 pKa = 4.74DD410 pKa = 4.67YY411 pKa = 12.15SSNLIVCKK419 pKa = 10.67ISDD422 pKa = 3.91TFNHH426 pKa = 5.92EE427 pKa = 4.2KK428 pKa = 10.77VSIEE432 pKa = 3.63AKK434 pKa = 10.15GIYY437 pKa = 9.66EE438 pKa = 4.69IEE440 pKa = 4.77DD441 pKa = 4.18EE442 pKa = 4.4DD443 pKa = 3.99WMIGPEE449 pKa = 3.83ITLAPNGEE457 pKa = 4.36FEE459 pKa = 4.55FSAEE463 pKa = 3.66ARR465 pKa = 11.84FFGGDD470 pKa = 3.16DD471 pKa = 4.1DD472 pKa = 5.2GRR474 pKa = 11.84LGQFDD479 pKa = 4.05EE480 pKa = 4.72SSYY483 pKa = 11.41LQFKK487 pKa = 8.9VKK489 pKa = 10.83AMFF492 pKa = 4.09
Molecular weight: 53.57 kDa
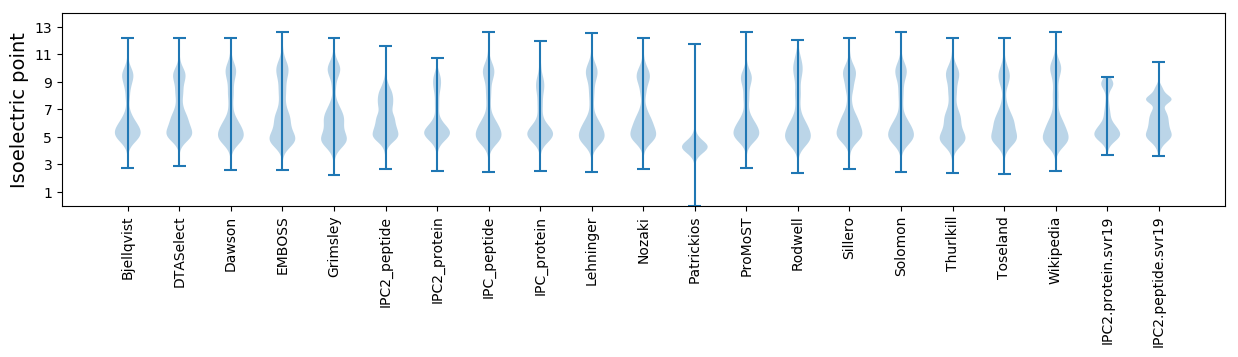
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1R3T1|E1R3T1_SEDSS O-acetylhomoserine/O-acetylserine sulfhydrylase OS=Sediminispirochaeta smaragdinae (strain DSM 11293 / JCM 15392 / SEBR 4228) OX=573413 GN=Spirs_2949 PE=3 SV=1
MM1 pKa = 7.49VKK3 pKa = 10.35LLCALGLVCLLSLLAGRR20 pKa = 11.84YY21 pKa = 8.01PSAGFTAPSTFIHH34 pKa = 7.29DD35 pKa = 3.61RR36 pKa = 11.84LARR39 pKa = 11.84NLLLSLRR46 pKa = 11.84LPRR49 pKa = 11.84LIAALLLGLALGAGGTVFQMLFSNPLVEE77 pKa = 4.68PGFLGVSQGAAFGAAAAIVLGGTAAWLIQISAACFAFVGLGASYY121 pKa = 11.27LLARR125 pKa = 11.84HH126 pKa = 5.71FRR128 pKa = 11.84FGGWTLRR135 pKa = 11.84LVLAGITVSALFSAGVGILKK155 pKa = 8.36YY156 pKa = 8.9TADD159 pKa = 4.23PMSQLPEE166 pKa = 3.59ITFWLLGGLWSVGWKK181 pKa = 9.62EE182 pKa = 4.08VLSVLPVVLLAALLLFAHH200 pKa = 7.43RR201 pKa = 11.84WRR203 pKa = 11.84LNLLSMDD210 pKa = 4.04DD211 pKa = 3.3RR212 pKa = 11.84TAFSFGIAPVRR223 pKa = 11.84EE224 pKa = 4.0RR225 pKa = 11.84LILLAAATAATAAVISISGMISWVGLIVPHH255 pKa = 6.06MARR258 pKa = 11.84KK259 pKa = 9.53LFSANSRR266 pKa = 11.84WSLPGAMMLGACFTIVCDD284 pKa = 3.68TLARR288 pKa = 11.84TLLAGEE294 pKa = 4.46IPLGVLTSFIGACFFIILLTKK315 pKa = 10.44KK316 pKa = 8.22STKK319 pKa = 9.91RR320 pKa = 11.84GRR322 pKa = 3.36
MM1 pKa = 7.49VKK3 pKa = 10.35LLCALGLVCLLSLLAGRR20 pKa = 11.84YY21 pKa = 8.01PSAGFTAPSTFIHH34 pKa = 7.29DD35 pKa = 3.61RR36 pKa = 11.84LARR39 pKa = 11.84NLLLSLRR46 pKa = 11.84LPRR49 pKa = 11.84LIAALLLGLALGAGGTVFQMLFSNPLVEE77 pKa = 4.68PGFLGVSQGAAFGAAAAIVLGGTAAWLIQISAACFAFVGLGASYY121 pKa = 11.27LLARR125 pKa = 11.84HH126 pKa = 5.71FRR128 pKa = 11.84FGGWTLRR135 pKa = 11.84LVLAGITVSALFSAGVGILKK155 pKa = 8.36YY156 pKa = 8.9TADD159 pKa = 4.23PMSQLPEE166 pKa = 3.59ITFWLLGGLWSVGWKK181 pKa = 9.62EE182 pKa = 4.08VLSVLPVVLLAALLLFAHH200 pKa = 7.43RR201 pKa = 11.84WRR203 pKa = 11.84LNLLSMDD210 pKa = 4.04DD211 pKa = 3.3RR212 pKa = 11.84TAFSFGIAPVRR223 pKa = 11.84EE224 pKa = 4.0RR225 pKa = 11.84LILLAAATAATAAVISISGMISWVGLIVPHH255 pKa = 6.06MARR258 pKa = 11.84KK259 pKa = 9.53LFSANSRR266 pKa = 11.84WSLPGAMMLGACFTIVCDD284 pKa = 3.68TLARR288 pKa = 11.84TLLAGEE294 pKa = 4.46IPLGVLTSFIGACFFIILLTKK315 pKa = 10.44KK316 pKa = 8.22STKK319 pKa = 9.91RR320 pKa = 11.84GRR322 pKa = 3.36
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1432067 |
31 |
3165 |
340.1 |
37.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.177 ± 0.042 | 1.026 ± 0.012 |
5.452 ± 0.032 | 7.064 ± 0.043 |
4.606 ± 0.024 | 7.657 ± 0.035 |
1.919 ± 0.017 | 7.154 ± 0.036 |
5.23 ± 0.033 | 10.287 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.575 ± 0.017 | 3.402 ± 0.024 |
4.092 ± 0.024 | 2.965 ± 0.02 |
5.718 ± 0.029 | 6.813 ± 0.031 |
4.956 ± 0.026 | 6.618 ± 0.03 |
1.038 ± 0.015 | 3.25 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
