
Planctomyces sp. SH-PL14
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Planctomyces; unclassified Planctomyces
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
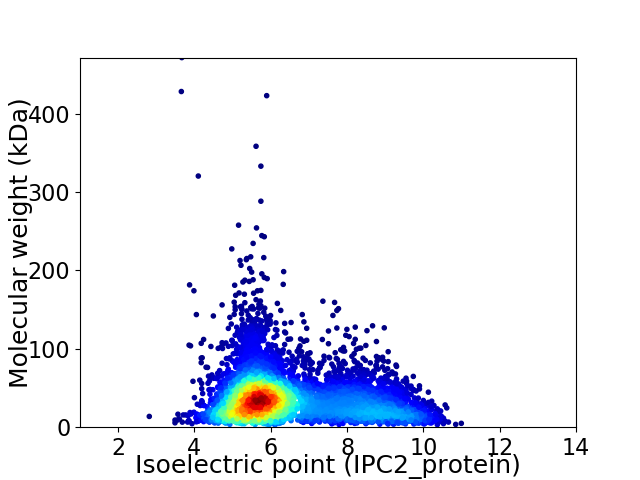
Virtual 2D-PAGE plot for 6497 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
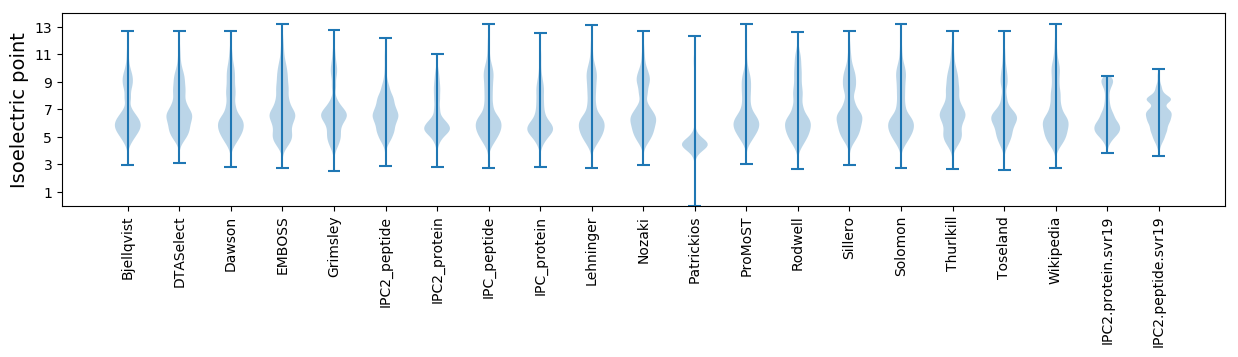
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142X417|A0A142X417_9PLAN Uncharacterized protein OS=Planctomyces sp. SH-PL14 OX=1632864 GN=VT03_27880 PE=4 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84YY3 pKa = 8.64FVAMAAVVTAVQLAAGVASAQDD25 pKa = 3.6LASPDD30 pKa = 3.58EE31 pKa = 4.16RR32 pKa = 11.84VEE34 pKa = 3.97MYY36 pKa = 10.51EE37 pKa = 4.1PEE39 pKa = 5.15DD40 pKa = 3.82YY41 pKa = 11.23ASLRR45 pKa = 11.84ARR47 pKa = 11.84IEE49 pKa = 3.8QLEE52 pKa = 3.99ANQRR56 pKa = 11.84YY57 pKa = 8.37SAPAVASLDD66 pKa = 3.87PQGGVYY72 pKa = 9.69VQYY75 pKa = 11.26DD76 pKa = 3.62NVIVKK81 pKa = 9.77PYY83 pKa = 10.2LSEE86 pKa = 4.15NSAFPAQIADD96 pKa = 3.8GNVVNYY102 pKa = 9.74GFNLDD107 pKa = 3.59MEE109 pKa = 4.55YY110 pKa = 10.71SPRR113 pKa = 11.84IEE115 pKa = 4.26VGYY118 pKa = 8.01MAPTSNLGFRR128 pKa = 11.84ARR130 pKa = 11.84YY131 pKa = 6.65WHH133 pKa = 6.72FGANAATGPANDD145 pKa = 4.34ANGLIADD152 pKa = 4.32DD153 pKa = 5.54AIAIAAGATIQDD165 pKa = 3.43VDD167 pKa = 4.08TITTVVYY174 pKa = 10.31DD175 pKa = 4.18LNMDD179 pKa = 3.93VLDD182 pKa = 4.65LEE184 pKa = 4.42ATRR187 pKa = 11.84AVGSSGTFAAGIRR200 pKa = 11.84TAWMGQKK207 pKa = 10.33YY208 pKa = 9.96GLDD211 pKa = 3.47SDD213 pKa = 4.17EE214 pKa = 4.81GQLFTDD220 pKa = 4.72LDD222 pKa = 3.87VAAIGPTLAYY232 pKa = 9.0EE233 pKa = 4.45WKK235 pKa = 10.46SHH237 pKa = 6.56DD238 pKa = 3.35FWFDD242 pKa = 3.12GLRR245 pKa = 11.84GLATVRR251 pKa = 11.84GSLLYY256 pKa = 10.49GGKK259 pKa = 10.08DD260 pKa = 3.28INSVGPDD267 pKa = 2.95NTYY270 pKa = 10.26TYY272 pKa = 10.89NEE274 pKa = 3.79QSLISNADD282 pKa = 3.31LQLGLEE288 pKa = 4.27YY289 pKa = 10.29RR290 pKa = 11.84RR291 pKa = 11.84SGGFFLRR298 pKa = 11.84AALEE302 pKa = 3.92AQYY305 pKa = 9.63WANTGNATNHH315 pKa = 5.54NVGPGDD321 pKa = 3.86DD322 pKa = 3.91VGDD325 pKa = 4.07SEE327 pKa = 5.08FVGSILDD334 pKa = 4.11DD335 pKa = 4.15DD336 pKa = 5.07LGFLGMVFSAGYY348 pKa = 9.32SWW350 pKa = 3.81
MM1 pKa = 7.91RR2 pKa = 11.84YY3 pKa = 8.64FVAMAAVVTAVQLAAGVASAQDD25 pKa = 3.6LASPDD30 pKa = 3.58EE31 pKa = 4.16RR32 pKa = 11.84VEE34 pKa = 3.97MYY36 pKa = 10.51EE37 pKa = 4.1PEE39 pKa = 5.15DD40 pKa = 3.82YY41 pKa = 11.23ASLRR45 pKa = 11.84ARR47 pKa = 11.84IEE49 pKa = 3.8QLEE52 pKa = 3.99ANQRR56 pKa = 11.84YY57 pKa = 8.37SAPAVASLDD66 pKa = 3.87PQGGVYY72 pKa = 9.69VQYY75 pKa = 11.26DD76 pKa = 3.62NVIVKK81 pKa = 9.77PYY83 pKa = 10.2LSEE86 pKa = 4.15NSAFPAQIADD96 pKa = 3.8GNVVNYY102 pKa = 9.74GFNLDD107 pKa = 3.59MEE109 pKa = 4.55YY110 pKa = 10.71SPRR113 pKa = 11.84IEE115 pKa = 4.26VGYY118 pKa = 8.01MAPTSNLGFRR128 pKa = 11.84ARR130 pKa = 11.84YY131 pKa = 6.65WHH133 pKa = 6.72FGANAATGPANDD145 pKa = 4.34ANGLIADD152 pKa = 4.32DD153 pKa = 5.54AIAIAAGATIQDD165 pKa = 3.43VDD167 pKa = 4.08TITTVVYY174 pKa = 10.31DD175 pKa = 4.18LNMDD179 pKa = 3.93VLDD182 pKa = 4.65LEE184 pKa = 4.42ATRR187 pKa = 11.84AVGSSGTFAAGIRR200 pKa = 11.84TAWMGQKK207 pKa = 10.33YY208 pKa = 9.96GLDD211 pKa = 3.47SDD213 pKa = 4.17EE214 pKa = 4.81GQLFTDD220 pKa = 4.72LDD222 pKa = 3.87VAAIGPTLAYY232 pKa = 9.0EE233 pKa = 4.45WKK235 pKa = 10.46SHH237 pKa = 6.56DD238 pKa = 3.35FWFDD242 pKa = 3.12GLRR245 pKa = 11.84GLATVRR251 pKa = 11.84GSLLYY256 pKa = 10.49GGKK259 pKa = 10.08DD260 pKa = 3.28INSVGPDD267 pKa = 2.95NTYY270 pKa = 10.26TYY272 pKa = 10.89NEE274 pKa = 3.79QSLISNADD282 pKa = 3.31LQLGLEE288 pKa = 4.27YY289 pKa = 10.29RR290 pKa = 11.84RR291 pKa = 11.84SGGFFLRR298 pKa = 11.84AALEE302 pKa = 3.92AQYY305 pKa = 9.63WANTGNATNHH315 pKa = 5.54NVGPGDD321 pKa = 3.86DD322 pKa = 3.91VGDD325 pKa = 4.07SEE327 pKa = 5.08FVGSILDD334 pKa = 4.11DD335 pKa = 4.15DD336 pKa = 5.07LGFLGMVFSAGYY348 pKa = 9.32SWW350 pKa = 3.81
Molecular weight: 37.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142X5Y7|A0A142X5Y7_9PLAN Circadian clock protein KaiB OS=Planctomyces sp. SH-PL14 OX=1632864 GN=kaiB PE=4 SV=1
MM1 pKa = 7.66SKK3 pKa = 10.55RR4 pKa = 11.84SGGPARR10 pKa = 11.84RR11 pKa = 11.84PATIKK16 pKa = 10.79PNLQKK21 pKa = 11.03KK22 pKa = 8.34MRR24 pKa = 11.84SQLRR28 pKa = 11.84PLKK31 pKa = 9.58TVSSKK36 pKa = 11.08SKK38 pKa = 9.91SGKK41 pKa = 9.78SGTRR45 pKa = 3.18
MM1 pKa = 7.66SKK3 pKa = 10.55RR4 pKa = 11.84SGGPARR10 pKa = 11.84RR11 pKa = 11.84PATIKK16 pKa = 10.79PNLQKK21 pKa = 11.03KK22 pKa = 8.34MRR24 pKa = 11.84SQLRR28 pKa = 11.84PLKK31 pKa = 9.58TVSSKK36 pKa = 11.08SKK38 pKa = 9.91SGKK41 pKa = 9.78SGTRR45 pKa = 3.18
Molecular weight: 4.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2309488 |
29 |
4696 |
355.5 |
38.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.74 ± 0.033 | 1.084 ± 0.012 |
5.602 ± 0.023 | 6.256 ± 0.025 |
3.606 ± 0.015 | 8.226 ± 0.032 |
2.109 ± 0.016 | 4.445 ± 0.022 |
3.442 ± 0.027 | 10.082 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.024 ± 0.013 | 2.608 ± 0.019 |
6.05 ± 0.032 | 3.584 ± 0.019 |
7.555 ± 0.037 | 5.831 ± 0.022 |
5.541 ± 0.031 | 7.381 ± 0.025 |
1.609 ± 0.015 | 2.225 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
