
Rhizobium mesoamericanum STM3625
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; Rhizobium mesoamericanum
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
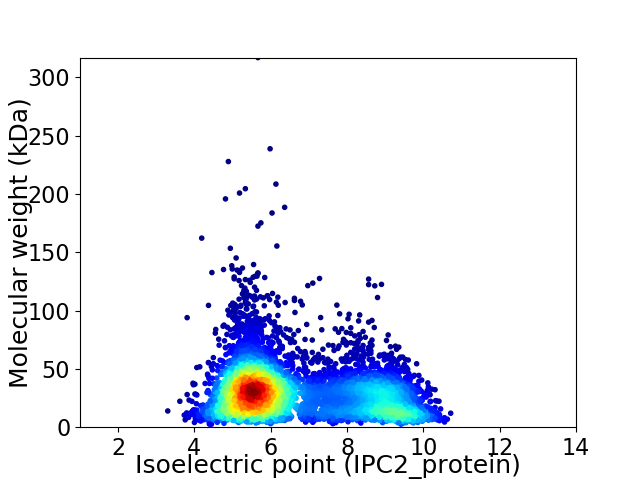
Virtual 2D-PAGE plot for 6358 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0PV03|K0PV03_9RHIZ Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex OS=Rhizobium mesoamericanum STM3625 OX=1211777 GN=sucB PE=3 SV=1
MM1 pKa = 6.91FTLAHH6 pKa = 6.81GGTITLPQGQTSVTVDD22 pKa = 2.87LHH24 pKa = 6.46FSADD28 pKa = 3.63TYY30 pKa = 11.22SGSISNSITSVTGQNEE46 pKa = 4.44YY47 pKa = 11.06EE48 pKa = 4.27SLLTTGSTSTTVNTPPTVVDD68 pKa = 3.67SEE70 pKa = 4.54NWLSSDD76 pKa = 3.31PAQYY80 pKa = 10.68TSGASSYY87 pKa = 10.26PNGYY91 pKa = 8.2PLHH94 pKa = 7.81IEE96 pKa = 4.24APTDD100 pKa = 3.48SDD102 pKa = 3.96HH103 pKa = 6.9NNLVVKK109 pKa = 10.61VSGTVPTGVFYY120 pKa = 11.15NGGSGYY126 pKa = 10.46VALTAGVTLYY136 pKa = 11.13DD137 pKa = 3.81LAHH140 pKa = 7.31GINLLDD146 pKa = 3.67SVVYY150 pKa = 10.4RR151 pKa = 11.84PTSSQTDD158 pKa = 3.61AQNLTLKK165 pKa = 10.62LDD167 pKa = 3.84VYY169 pKa = 11.15DD170 pKa = 3.88GTVHH174 pKa = 5.14VTQNVGIHH182 pKa = 5.47EE183 pKa = 4.65VPPTSLPSDD192 pKa = 3.74VQQIGSGGSPLTSGNDD208 pKa = 3.24QTQTLTLSQGTVNGILADD226 pKa = 3.76PHH228 pKa = 6.37GSTVVVYY235 pKa = 10.5TDD237 pKa = 3.81FQKK240 pKa = 11.21SPFATPIPADD250 pKa = 3.74EE251 pKa = 4.21QNPGAFGDD259 pKa = 4.63GSAGSARR266 pKa = 11.84EE267 pKa = 3.67QEE269 pKa = 4.45VQVEE273 pKa = 3.95IRR275 pKa = 11.84IGANRR280 pKa = 11.84FAVVEE285 pKa = 4.27DD286 pKa = 4.18DD287 pKa = 3.88RR288 pKa = 11.84SAGTFEE294 pKa = 4.37QSWFYY299 pKa = 11.63DD300 pKa = 3.65SASGLMKK307 pKa = 10.58AAVDD311 pKa = 3.6YY312 pKa = 11.35DD313 pKa = 4.1HH314 pKa = 7.63IYY316 pKa = 11.06LLDD319 pKa = 3.88AAGNATSTTLASYY332 pKa = 10.66LIGNPPTAGNTWTLVYY348 pKa = 10.29TDD350 pKa = 3.93NNGGNFQARR359 pKa = 11.84TVTFDD364 pKa = 4.1FFTHH368 pKa = 6.19NPGDD372 pKa = 3.55PGIPVTGNTTLADD385 pKa = 4.26TIYY388 pKa = 8.92GTSGEE393 pKa = 4.34DD394 pKa = 3.26VLTGSGGNDD403 pKa = 3.14MLYY406 pKa = 10.72GRR408 pKa = 11.84QDD410 pKa = 3.03HH411 pKa = 7.6DD412 pKa = 3.84ILNGGIGNDD421 pKa = 3.69MLTGGADD428 pKa = 3.6ADD430 pKa = 4.29TLTGGSGADD439 pKa = 3.2TFVIGSGEE447 pKa = 4.23SLGTTGGNGDD457 pKa = 3.9NGTISGNDD465 pKa = 3.39IITDD469 pKa = 4.44FDD471 pKa = 3.66TTADD475 pKa = 3.56ILNLAGTPFAAGNTSGWVDD494 pKa = 3.5GTNSSLTISSSQISKK509 pKa = 10.71HH510 pKa = 6.15SITNGIITFEE520 pKa = 4.03NASNNVVTLSSASQVAAVVQYY541 pKa = 9.76LHH543 pKa = 6.41QNDD546 pKa = 3.87MGNAGATVAFTATIGGTAHH565 pKa = 6.2TFIYY569 pKa = 9.44EE570 pKa = 4.25QVGDD574 pKa = 4.11TPDD577 pKa = 3.09ASKK580 pKa = 11.1DD581 pKa = 3.41ILVDD585 pKa = 3.94LQNVNLISGGTSLATLIGNSHH606 pKa = 7.33IDD608 pKa = 4.19PIVLDD613 pKa = 4.05LDD615 pKa = 4.16HH616 pKa = 7.07NGVALTSLDD625 pKa = 3.24QGVQFDD631 pKa = 4.25INADD635 pKa = 3.44GHH637 pKa = 6.51KK638 pKa = 10.7DD639 pKa = 3.44QIAWTTGSDD648 pKa = 3.69GILAFDD654 pKa = 3.48VDD656 pKa = 4.57GNGKK660 pKa = 9.03IDD662 pKa = 3.59NGSEE666 pKa = 3.55IFSPHH671 pKa = 5.93FAGGSYY677 pKa = 10.96VDD679 pKa = 4.32GLAALSALDD688 pKa = 4.14SNHH691 pKa = 7.05DD692 pKa = 3.82GKK694 pKa = 10.68IDD696 pKa = 3.64AADD699 pKa = 3.5EE700 pKa = 4.48AFSKK704 pKa = 9.35LTVWQDD710 pKa = 3.31LNHH713 pKa = 6.66NGITDD718 pKa = 3.61SGEE721 pKa = 4.0LSSLADD727 pKa = 3.63HH728 pKa = 6.91SISSISLEE736 pKa = 3.9ATASSTEE743 pKa = 3.85INGQSILADD752 pKa = 3.42GDD754 pKa = 3.87YY755 pKa = 10.65TLTDD759 pKa = 3.32GSTAHH764 pKa = 5.71FVEE767 pKa = 4.88VAFDD771 pKa = 3.59TTLGGSEE778 pKa = 4.12NSSNAYY784 pKa = 10.36SLIGSDD790 pKa = 4.51GDD792 pKa = 4.93DD793 pKa = 3.37ILSGSGGMFTISGGAGADD811 pKa = 3.66TFVLDD816 pKa = 4.84ADD818 pKa = 4.06ALNDD822 pKa = 3.48VKK824 pKa = 10.93LADD827 pKa = 4.12VITDD831 pKa = 4.01FKK833 pKa = 11.34ASEE836 pKa = 4.42GDD838 pKa = 3.46TLDD841 pKa = 3.29VSKK844 pKa = 11.07LLDD847 pKa = 3.59SLLGHH852 pKa = 5.88QASEE856 pKa = 4.47AEE858 pKa = 4.11ALASVKK864 pKa = 8.25TTVSGADD871 pKa = 3.11TVVSVNANGGWHH883 pKa = 6.85DD884 pKa = 3.83VAVLQNTTEE893 pKa = 3.9AVKK896 pKa = 10.53ILFDD900 pKa = 4.8DD901 pKa = 3.81KK902 pKa = 11.36HH903 pKa = 6.29DD904 pKa = 3.83TTTAPHH910 pKa = 5.91VGG912 pKa = 3.39
MM1 pKa = 6.91FTLAHH6 pKa = 6.81GGTITLPQGQTSVTVDD22 pKa = 2.87LHH24 pKa = 6.46FSADD28 pKa = 3.63TYY30 pKa = 11.22SGSISNSITSVTGQNEE46 pKa = 4.44YY47 pKa = 11.06EE48 pKa = 4.27SLLTTGSTSTTVNTPPTVVDD68 pKa = 3.67SEE70 pKa = 4.54NWLSSDD76 pKa = 3.31PAQYY80 pKa = 10.68TSGASSYY87 pKa = 10.26PNGYY91 pKa = 8.2PLHH94 pKa = 7.81IEE96 pKa = 4.24APTDD100 pKa = 3.48SDD102 pKa = 3.96HH103 pKa = 6.9NNLVVKK109 pKa = 10.61VSGTVPTGVFYY120 pKa = 11.15NGGSGYY126 pKa = 10.46VALTAGVTLYY136 pKa = 11.13DD137 pKa = 3.81LAHH140 pKa = 7.31GINLLDD146 pKa = 3.67SVVYY150 pKa = 10.4RR151 pKa = 11.84PTSSQTDD158 pKa = 3.61AQNLTLKK165 pKa = 10.62LDD167 pKa = 3.84VYY169 pKa = 11.15DD170 pKa = 3.88GTVHH174 pKa = 5.14VTQNVGIHH182 pKa = 5.47EE183 pKa = 4.65VPPTSLPSDD192 pKa = 3.74VQQIGSGGSPLTSGNDD208 pKa = 3.24QTQTLTLSQGTVNGILADD226 pKa = 3.76PHH228 pKa = 6.37GSTVVVYY235 pKa = 10.5TDD237 pKa = 3.81FQKK240 pKa = 11.21SPFATPIPADD250 pKa = 3.74EE251 pKa = 4.21QNPGAFGDD259 pKa = 4.63GSAGSARR266 pKa = 11.84EE267 pKa = 3.67QEE269 pKa = 4.45VQVEE273 pKa = 3.95IRR275 pKa = 11.84IGANRR280 pKa = 11.84FAVVEE285 pKa = 4.27DD286 pKa = 4.18DD287 pKa = 3.88RR288 pKa = 11.84SAGTFEE294 pKa = 4.37QSWFYY299 pKa = 11.63DD300 pKa = 3.65SASGLMKK307 pKa = 10.58AAVDD311 pKa = 3.6YY312 pKa = 11.35DD313 pKa = 4.1HH314 pKa = 7.63IYY316 pKa = 11.06LLDD319 pKa = 3.88AAGNATSTTLASYY332 pKa = 10.66LIGNPPTAGNTWTLVYY348 pKa = 10.29TDD350 pKa = 3.93NNGGNFQARR359 pKa = 11.84TVTFDD364 pKa = 4.1FFTHH368 pKa = 6.19NPGDD372 pKa = 3.55PGIPVTGNTTLADD385 pKa = 4.26TIYY388 pKa = 8.92GTSGEE393 pKa = 4.34DD394 pKa = 3.26VLTGSGGNDD403 pKa = 3.14MLYY406 pKa = 10.72GRR408 pKa = 11.84QDD410 pKa = 3.03HH411 pKa = 7.6DD412 pKa = 3.84ILNGGIGNDD421 pKa = 3.69MLTGGADD428 pKa = 3.6ADD430 pKa = 4.29TLTGGSGADD439 pKa = 3.2TFVIGSGEE447 pKa = 4.23SLGTTGGNGDD457 pKa = 3.9NGTISGNDD465 pKa = 3.39IITDD469 pKa = 4.44FDD471 pKa = 3.66TTADD475 pKa = 3.56ILNLAGTPFAAGNTSGWVDD494 pKa = 3.5GTNSSLTISSSQISKK509 pKa = 10.71HH510 pKa = 6.15SITNGIITFEE520 pKa = 4.03NASNNVVTLSSASQVAAVVQYY541 pKa = 9.76LHH543 pKa = 6.41QNDD546 pKa = 3.87MGNAGATVAFTATIGGTAHH565 pKa = 6.2TFIYY569 pKa = 9.44EE570 pKa = 4.25QVGDD574 pKa = 4.11TPDD577 pKa = 3.09ASKK580 pKa = 11.1DD581 pKa = 3.41ILVDD585 pKa = 3.94LQNVNLISGGTSLATLIGNSHH606 pKa = 7.33IDD608 pKa = 4.19PIVLDD613 pKa = 4.05LDD615 pKa = 4.16HH616 pKa = 7.07NGVALTSLDD625 pKa = 3.24QGVQFDD631 pKa = 4.25INADD635 pKa = 3.44GHH637 pKa = 6.51KK638 pKa = 10.7DD639 pKa = 3.44QIAWTTGSDD648 pKa = 3.69GILAFDD654 pKa = 3.48VDD656 pKa = 4.57GNGKK660 pKa = 9.03IDD662 pKa = 3.59NGSEE666 pKa = 3.55IFSPHH671 pKa = 5.93FAGGSYY677 pKa = 10.96VDD679 pKa = 4.32GLAALSALDD688 pKa = 4.14SNHH691 pKa = 7.05DD692 pKa = 3.82GKK694 pKa = 10.68IDD696 pKa = 3.64AADD699 pKa = 3.5EE700 pKa = 4.48AFSKK704 pKa = 9.35LTVWQDD710 pKa = 3.31LNHH713 pKa = 6.66NGITDD718 pKa = 3.61SGEE721 pKa = 4.0LSSLADD727 pKa = 3.63HH728 pKa = 6.91SISSISLEE736 pKa = 3.9ATASSTEE743 pKa = 3.85INGQSILADD752 pKa = 3.42GDD754 pKa = 3.87YY755 pKa = 10.65TLTDD759 pKa = 3.32GSTAHH764 pKa = 5.71FVEE767 pKa = 4.88VAFDD771 pKa = 3.59TTLGGSEE778 pKa = 4.12NSSNAYY784 pKa = 10.36SLIGSDD790 pKa = 4.51GDD792 pKa = 4.93DD793 pKa = 3.37ILSGSGGMFTISGGAGADD811 pKa = 3.66TFVLDD816 pKa = 4.84ADD818 pKa = 4.06ALNDD822 pKa = 3.48VKK824 pKa = 10.93LADD827 pKa = 4.12VITDD831 pKa = 4.01FKK833 pKa = 11.34ASEE836 pKa = 4.42GDD838 pKa = 3.46TLDD841 pKa = 3.29VSKK844 pKa = 11.07LLDD847 pKa = 3.59SLLGHH852 pKa = 5.88QASEE856 pKa = 4.47AEE858 pKa = 4.11ALASVKK864 pKa = 8.25TTVSGADD871 pKa = 3.11TVVSVNANGGWHH883 pKa = 6.85DD884 pKa = 3.83VAVLQNTTEE893 pKa = 3.9AVKK896 pKa = 10.53ILFDD900 pKa = 4.8DD901 pKa = 3.81KK902 pKa = 11.36HH903 pKa = 6.29DD904 pKa = 3.83TTTAPHH910 pKa = 5.91VGG912 pKa = 3.39
Molecular weight: 93.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0PYX2|K0PYX2_9RHIZ D-alanine--D-alanine ligase OS=Rhizobium mesoamericanum STM3625 OX=1211777 GN=ddlB PE=3 SV=1
MM1 pKa = 7.7DD2 pKa = 4.3TPAFAASVVALLILPGPTNAVLAMASHH29 pKa = 7.05GLRR32 pKa = 11.84LSRR35 pKa = 11.84MGVLLVAVILTYY47 pKa = 10.72LAVIVPVSLFGGPFLQQHH65 pKa = 6.29PAVSQGVKK73 pKa = 9.72LASATWMFYY82 pKa = 10.79LALKK86 pKa = 10.05LWGRR90 pKa = 11.84PSRR93 pKa = 11.84QMQPVAIGVRR103 pKa = 11.84QVVATTLLNPKK114 pKa = 10.11AIIIALAMLPSRR126 pKa = 11.84GILAPDD132 pKa = 4.02LFVSFVVAAGGASCMWLSIGRR153 pKa = 11.84LVLGEE158 pKa = 4.21GDD160 pKa = 4.05QMPLAARR167 pKa = 11.84RR168 pKa = 11.84CGSAVLLGFCVLLALSALL186 pKa = 4.0
MM1 pKa = 7.7DD2 pKa = 4.3TPAFAASVVALLILPGPTNAVLAMASHH29 pKa = 7.05GLRR32 pKa = 11.84LSRR35 pKa = 11.84MGVLLVAVILTYY47 pKa = 10.72LAVIVPVSLFGGPFLQQHH65 pKa = 6.29PAVSQGVKK73 pKa = 9.72LASATWMFYY82 pKa = 10.79LALKK86 pKa = 10.05LWGRR90 pKa = 11.84PSRR93 pKa = 11.84QMQPVAIGVRR103 pKa = 11.84QVVATTLLNPKK114 pKa = 10.11AIIIALAMLPSRR126 pKa = 11.84GILAPDD132 pKa = 4.02LFVSFVVAAGGASCMWLSIGRR153 pKa = 11.84LVLGEE158 pKa = 4.21GDD160 pKa = 4.05QMPLAARR167 pKa = 11.84RR168 pKa = 11.84CGSAVLLGFCVLLALSALL186 pKa = 4.0
Molecular weight: 19.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1835771 |
21 |
2839 |
288.7 |
31.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.614 ± 0.042 | 0.876 ± 0.011 |
5.593 ± 0.024 | 5.703 ± 0.029 |
3.956 ± 0.023 | 8.112 ± 0.027 |
2.105 ± 0.015 | 5.748 ± 0.025 |
3.826 ± 0.024 | 9.947 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.015 | 2.934 ± 0.016 |
4.837 ± 0.02 | 3.164 ± 0.017 |
6.745 ± 0.032 | 5.978 ± 0.024 |
5.328 ± 0.022 | 7.336 ± 0.021 |
1.307 ± 0.015 | 2.357 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
