
Streptococcus satellite phage Javan217
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
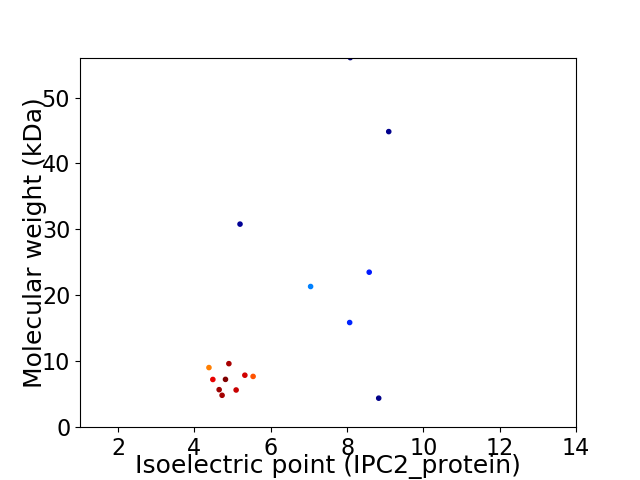
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
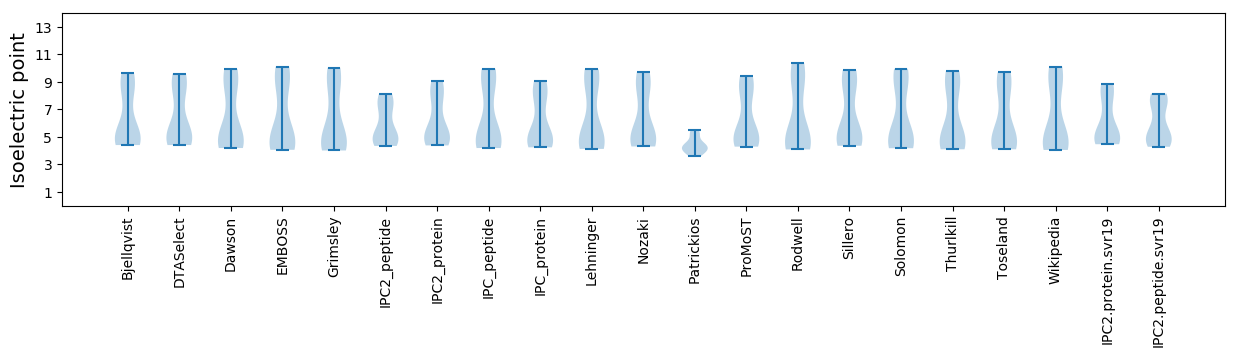
* You can choose from 21 different methods for calculating isoelectric point
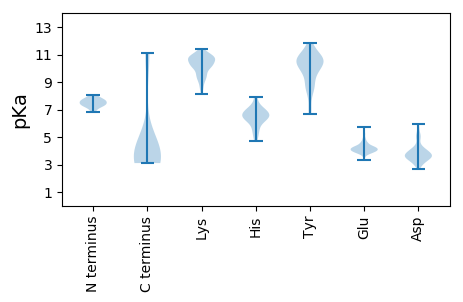
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZGE6|A0A4D5ZGE6_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan217 OX=2558574 GN=JavanS217_0004 PE=4 SV=1
MM1 pKa = 7.57LVRR4 pKa = 11.84LEE6 pKa = 4.24FSDD9 pKa = 4.86GNTLTRR15 pKa = 11.84MVSKK19 pKa = 9.15HH20 pKa = 5.01TLDD23 pKa = 4.03MILARR28 pKa = 11.84PYY30 pKa = 10.92SEE32 pKa = 3.92TWLHH36 pKa = 6.67FNGEE40 pKa = 4.28KK41 pKa = 10.04IAVLEE46 pKa = 4.01NVEE49 pKa = 4.1VLGNVDD55 pKa = 4.92DD56 pKa = 5.97FDD58 pKa = 5.52NEE60 pKa = 4.21VNKK63 pKa = 10.09TIAYY67 pKa = 9.79NEE69 pKa = 4.4LEE71 pKa = 4.3NEE73 pKa = 4.27VQEE76 pKa = 4.56NEE78 pKa = 3.94
MM1 pKa = 7.57LVRR4 pKa = 11.84LEE6 pKa = 4.24FSDD9 pKa = 4.86GNTLTRR15 pKa = 11.84MVSKK19 pKa = 9.15HH20 pKa = 5.01TLDD23 pKa = 4.03MILARR28 pKa = 11.84PYY30 pKa = 10.92SEE32 pKa = 3.92TWLHH36 pKa = 6.67FNGEE40 pKa = 4.28KK41 pKa = 10.04IAVLEE46 pKa = 4.01NVEE49 pKa = 4.1VLGNVDD55 pKa = 4.92DD56 pKa = 5.97FDD58 pKa = 5.52NEE60 pKa = 4.21VNKK63 pKa = 10.09TIAYY67 pKa = 9.79NEE69 pKa = 4.4LEE71 pKa = 4.3NEE73 pKa = 4.27VQEE76 pKa = 4.56NEE78 pKa = 3.94
Molecular weight: 9.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZGP8|A0A4D5ZGP8_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan217 OX=2558574 GN=JavanS217_0011 PE=4 SV=1
MM1 pKa = 8.09AEE3 pKa = 3.91IKK5 pKa = 10.55KK6 pKa = 8.29ITKK9 pKa = 10.26KK10 pKa = 10.74NGTTVYY16 pKa = 10.04RR17 pKa = 11.84QQIYY21 pKa = 10.8LGTDD25 pKa = 3.14CMTGKK30 pKa = 9.72QVYY33 pKa = 8.65TSISAPTKK41 pKa = 10.55KK42 pKa = 9.92EE43 pKa = 3.83LKK45 pKa = 9.55QKK47 pKa = 10.85RR48 pKa = 11.84EE49 pKa = 3.75FKK51 pKa = 10.77INEE54 pKa = 4.18FKK56 pKa = 11.23DD57 pKa = 3.31NGYY60 pKa = 7.94TRR62 pKa = 11.84YY63 pKa = 10.57KK64 pKa = 10.47SVTVKK69 pKa = 10.57NYY71 pKa = 10.67RR72 pKa = 11.84EE73 pKa = 4.16LSEE76 pKa = 4.8LWLEE80 pKa = 4.02NHH82 pKa = 6.29KK83 pKa = 11.28LEE85 pKa = 5.31VKK87 pKa = 9.27PQSYY91 pKa = 10.89KK92 pKa = 9.75NTIIKK97 pKa = 9.6LNKK100 pKa = 9.2YY101 pKa = 8.6LLPVFGDD108 pKa = 3.35MKK110 pKa = 10.73VEE112 pKa = 4.57KK113 pKa = 9.09ITLPMVQNFVNKK125 pKa = 10.19LVGSEE130 pKa = 3.89LTSYY134 pKa = 10.87KK135 pKa = 10.58VVISINKK142 pKa = 9.65RR143 pKa = 11.84ILKK146 pKa = 9.77YY147 pKa = 10.46AVNLQIINVNPADD160 pKa = 3.76NVIVPKK166 pKa = 10.52VKK168 pKa = 10.1KK169 pKa = 10.12SKK171 pKa = 10.2KK172 pKa = 9.04EE173 pKa = 3.93KK174 pKa = 10.31VKK176 pKa = 10.98HH177 pKa = 6.29FDD179 pKa = 4.08DD180 pKa = 4.37SQLKK184 pKa = 8.91QFKK187 pKa = 10.52DD188 pKa = 3.78YY189 pKa = 10.76MNSLSDD195 pKa = 3.42TFRR198 pKa = 11.84NCYY201 pKa = 10.17YY202 pKa = 9.33KK203 pKa = 10.01TLYY206 pKa = 8.72YY207 pKa = 10.26TLLASGLRR215 pKa = 11.84IGEE218 pKa = 4.49AIALEE223 pKa = 4.01WSDD226 pKa = 5.15IYY228 pKa = 11.51LEE230 pKa = 4.95DD231 pKa = 5.18LDD233 pKa = 4.92NGYY236 pKa = 10.04IDD238 pKa = 3.62VNKK241 pKa = 10.13TYY243 pKa = 10.68VEE245 pKa = 4.16KK246 pKa = 10.8EE247 pKa = 3.85KK248 pKa = 11.03GVNDD252 pKa = 4.14PKK254 pKa = 11.08SEE256 pKa = 3.94AGKK259 pKa = 10.28RR260 pKa = 11.84RR261 pKa = 11.84VPIDD265 pKa = 3.41RR266 pKa = 11.84NTVLMLRR273 pKa = 11.84LYY275 pKa = 10.42KK276 pKa = 10.33ARR278 pKa = 11.84QRR280 pKa = 11.84QCFMLNGYY288 pKa = 7.03GAKK291 pKa = 8.14MTEE294 pKa = 4.21HH295 pKa = 6.15VFSNGFSAYY304 pKa = 6.65PTTVGLQYY312 pKa = 11.56VLTQHH317 pKa = 6.82LQQAGLPRR325 pKa = 11.84FTFHH329 pKa = 7.9AFRR332 pKa = 11.84HH333 pKa = 4.69THH335 pKa = 6.92ASLLLNAGIGYY346 pKa = 9.74KK347 pKa = 9.96EE348 pKa = 3.9LQHH351 pKa = 6.77RR352 pKa = 11.84LGHH355 pKa = 5.56STLAMTMDD363 pKa = 4.54TYY365 pKa = 11.85SHH367 pKa = 7.05LSKK370 pKa = 10.7EE371 pKa = 4.25KK372 pKa = 10.83EE373 pKa = 3.53KK374 pKa = 11.22DD375 pKa = 3.16AVNFFEE381 pKa = 5.73KK382 pKa = 10.83AMANLL387 pKa = 3.66
MM1 pKa = 8.09AEE3 pKa = 3.91IKK5 pKa = 10.55KK6 pKa = 8.29ITKK9 pKa = 10.26KK10 pKa = 10.74NGTTVYY16 pKa = 10.04RR17 pKa = 11.84QQIYY21 pKa = 10.8LGTDD25 pKa = 3.14CMTGKK30 pKa = 9.72QVYY33 pKa = 8.65TSISAPTKK41 pKa = 10.55KK42 pKa = 9.92EE43 pKa = 3.83LKK45 pKa = 9.55QKK47 pKa = 10.85RR48 pKa = 11.84EE49 pKa = 3.75FKK51 pKa = 10.77INEE54 pKa = 4.18FKK56 pKa = 11.23DD57 pKa = 3.31NGYY60 pKa = 7.94TRR62 pKa = 11.84YY63 pKa = 10.57KK64 pKa = 10.47SVTVKK69 pKa = 10.57NYY71 pKa = 10.67RR72 pKa = 11.84EE73 pKa = 4.16LSEE76 pKa = 4.8LWLEE80 pKa = 4.02NHH82 pKa = 6.29KK83 pKa = 11.28LEE85 pKa = 5.31VKK87 pKa = 9.27PQSYY91 pKa = 10.89KK92 pKa = 9.75NTIIKK97 pKa = 9.6LNKK100 pKa = 9.2YY101 pKa = 8.6LLPVFGDD108 pKa = 3.35MKK110 pKa = 10.73VEE112 pKa = 4.57KK113 pKa = 9.09ITLPMVQNFVNKK125 pKa = 10.19LVGSEE130 pKa = 3.89LTSYY134 pKa = 10.87KK135 pKa = 10.58VVISINKK142 pKa = 9.65RR143 pKa = 11.84ILKK146 pKa = 9.77YY147 pKa = 10.46AVNLQIINVNPADD160 pKa = 3.76NVIVPKK166 pKa = 10.52VKK168 pKa = 10.1KK169 pKa = 10.12SKK171 pKa = 10.2KK172 pKa = 9.04EE173 pKa = 3.93KK174 pKa = 10.31VKK176 pKa = 10.98HH177 pKa = 6.29FDD179 pKa = 4.08DD180 pKa = 4.37SQLKK184 pKa = 8.91QFKK187 pKa = 10.52DD188 pKa = 3.78YY189 pKa = 10.76MNSLSDD195 pKa = 3.42TFRR198 pKa = 11.84NCYY201 pKa = 10.17YY202 pKa = 9.33KK203 pKa = 10.01TLYY206 pKa = 8.72YY207 pKa = 10.26TLLASGLRR215 pKa = 11.84IGEE218 pKa = 4.49AIALEE223 pKa = 4.01WSDD226 pKa = 5.15IYY228 pKa = 11.51LEE230 pKa = 4.95DD231 pKa = 5.18LDD233 pKa = 4.92NGYY236 pKa = 10.04IDD238 pKa = 3.62VNKK241 pKa = 10.13TYY243 pKa = 10.68VEE245 pKa = 4.16KK246 pKa = 10.8EE247 pKa = 3.85KK248 pKa = 11.03GVNDD252 pKa = 4.14PKK254 pKa = 11.08SEE256 pKa = 3.94AGKK259 pKa = 10.28RR260 pKa = 11.84RR261 pKa = 11.84VPIDD265 pKa = 3.41RR266 pKa = 11.84NTVLMLRR273 pKa = 11.84LYY275 pKa = 10.42KK276 pKa = 10.33ARR278 pKa = 11.84QRR280 pKa = 11.84QCFMLNGYY288 pKa = 7.03GAKK291 pKa = 8.14MTEE294 pKa = 4.21HH295 pKa = 6.15VFSNGFSAYY304 pKa = 6.65PTTVGLQYY312 pKa = 11.56VLTQHH317 pKa = 6.82LQQAGLPRR325 pKa = 11.84FTFHH329 pKa = 7.9AFRR332 pKa = 11.84HH333 pKa = 4.69THH335 pKa = 6.92ASLLLNAGIGYY346 pKa = 9.74KK347 pKa = 9.96EE348 pKa = 3.9LQHH351 pKa = 6.77RR352 pKa = 11.84LGHH355 pKa = 5.56STLAMTMDD363 pKa = 4.54TYY365 pKa = 11.85SHH367 pKa = 7.05LSKK370 pKa = 10.7EE371 pKa = 4.25KK372 pKa = 10.83EE373 pKa = 3.53KK374 pKa = 11.22DD375 pKa = 3.16AVNFFEE381 pKa = 5.73KK382 pKa = 10.83AMANLL387 pKa = 3.66
Molecular weight: 44.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2255 |
37 |
476 |
140.9 |
16.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.922 ± 0.49 | 0.443 ± 0.108 |
6.519 ± 0.661 | 8.337 ± 0.739 |
3.725 ± 0.383 | 4.346 ± 0.36 |
1.552 ± 0.284 | 6.696 ± 0.559 |
10.067 ± 0.437 | 10.51 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.572 ± 0.367 | 6.519 ± 0.446 |
2.217 ± 0.256 | 3.681 ± 0.289 |
4.435 ± 0.511 | 5.011 ± 0.328 |
6.164 ± 0.356 | 6.519 ± 0.494 |
0.843 ± 0.15 | 4.922 ± 0.554 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
