
Psychromonas sp. CNPT3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Psychromonas; unclassified Psychromonas
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
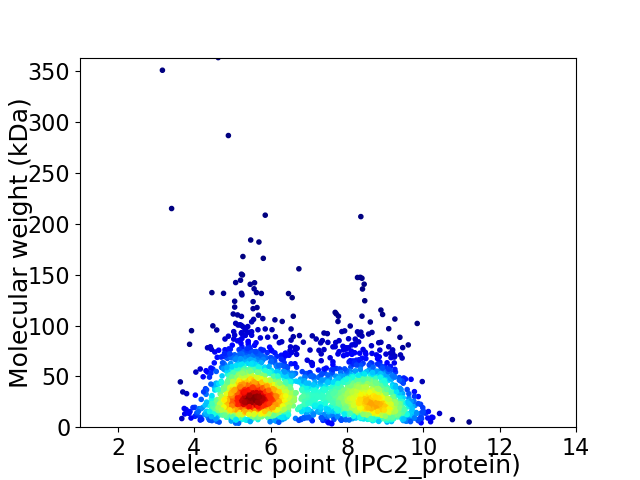
Virtual 2D-PAGE plot for 2578 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4UP76|M4UP76_9GAMM PTS system N-acetylgalactosamine-specific transporter subunit IID OS=Psychromonas sp. CNPT3 OX=314282 GN=PCNPT3_11895 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.43KK3 pKa = 9.06TILAMAITGLFAATAAQAATVYY25 pKa = 10.66DD26 pKa = 3.96ADD28 pKa = 4.42GVTLKK33 pKa = 11.18LNGDD37 pKa = 3.55VKK39 pKa = 10.83INYY42 pKa = 9.49GSDD45 pKa = 2.96VRR47 pKa = 11.84KK48 pKa = 10.17FNDD51 pKa = 3.04NKK53 pKa = 10.26VDD55 pKa = 3.6SNYY58 pKa = 9.86IEE60 pKa = 5.87LDD62 pKa = 3.48DD63 pKa = 5.59ADD65 pKa = 4.43FSFALGYY72 pKa = 10.51EE73 pKa = 3.9IGNGIEE79 pKa = 4.61LGATMEE85 pKa = 5.08ISGEE89 pKa = 3.99DD90 pKa = 3.64DD91 pKa = 4.43NIALSDD97 pKa = 3.7TFLSVAGDD105 pKa = 4.15FGTVTVGKK113 pKa = 10.08QPLIFDD119 pKa = 4.1DD120 pKa = 4.37AGIGEE125 pKa = 4.35DD126 pKa = 3.97FKK128 pKa = 11.62FGFDD132 pKa = 3.26TYY134 pKa = 11.89ANTTDD139 pKa = 4.02SGEE142 pKa = 3.92QVIKK146 pKa = 11.27YY147 pKa = 10.14KK148 pKa = 10.98GDD150 pKa = 3.05WDD152 pKa = 3.66TFYY155 pKa = 11.47AGVAYY160 pKa = 9.27MLNGDD165 pKa = 3.79ATSNGDD171 pKa = 3.21GTTGEE176 pKa = 4.14DD177 pKa = 4.87DD178 pKa = 4.21KK179 pKa = 11.91NQVDD183 pKa = 3.65ANFGVRR189 pKa = 11.84FAGIDD194 pKa = 3.26AAVYY198 pKa = 10.68YY199 pKa = 9.79STGTNNLKK207 pKa = 10.05QDD209 pKa = 3.33NNAYY213 pKa = 9.63ILQAMYY219 pKa = 10.56NADD222 pKa = 3.69AFKK225 pKa = 11.14VGAFYY230 pKa = 11.22SNEE233 pKa = 4.05TIEE236 pKa = 4.34NSNGSDD242 pKa = 3.69LSDD245 pKa = 3.73LNHH248 pKa = 6.41YY249 pKa = 8.88GVSGNYY255 pKa = 10.35YY256 pKa = 8.33MDD258 pKa = 3.05AWNFGLGWGAEE269 pKa = 3.97KK270 pKa = 11.12DD271 pKa = 3.64NLTSNNDD278 pKa = 3.39RR279 pKa = 11.84NDD281 pKa = 3.07YY282 pKa = 8.53YY283 pKa = 11.88ANVAYY288 pKa = 10.27AFTSNIQSYY297 pKa = 9.65VEE299 pKa = 3.9VGYY302 pKa = 10.91SDD304 pKa = 4.83EE305 pKa = 5.21DD306 pKa = 4.04DD307 pKa = 4.68SEE309 pKa = 4.34VGYY312 pKa = 10.56VVGMEE317 pKa = 4.24VVFF320 pKa = 4.62
MM1 pKa = 7.68KK2 pKa = 9.43KK3 pKa = 9.06TILAMAITGLFAATAAQAATVYY25 pKa = 10.66DD26 pKa = 3.96ADD28 pKa = 4.42GVTLKK33 pKa = 11.18LNGDD37 pKa = 3.55VKK39 pKa = 10.83INYY42 pKa = 9.49GSDD45 pKa = 2.96VRR47 pKa = 11.84KK48 pKa = 10.17FNDD51 pKa = 3.04NKK53 pKa = 10.26VDD55 pKa = 3.6SNYY58 pKa = 9.86IEE60 pKa = 5.87LDD62 pKa = 3.48DD63 pKa = 5.59ADD65 pKa = 4.43FSFALGYY72 pKa = 10.51EE73 pKa = 3.9IGNGIEE79 pKa = 4.61LGATMEE85 pKa = 5.08ISGEE89 pKa = 3.99DD90 pKa = 3.64DD91 pKa = 4.43NIALSDD97 pKa = 3.7TFLSVAGDD105 pKa = 4.15FGTVTVGKK113 pKa = 10.08QPLIFDD119 pKa = 4.1DD120 pKa = 4.37AGIGEE125 pKa = 4.35DD126 pKa = 3.97FKK128 pKa = 11.62FGFDD132 pKa = 3.26TYY134 pKa = 11.89ANTTDD139 pKa = 4.02SGEE142 pKa = 3.92QVIKK146 pKa = 11.27YY147 pKa = 10.14KK148 pKa = 10.98GDD150 pKa = 3.05WDD152 pKa = 3.66TFYY155 pKa = 11.47AGVAYY160 pKa = 9.27MLNGDD165 pKa = 3.79ATSNGDD171 pKa = 3.21GTTGEE176 pKa = 4.14DD177 pKa = 4.87DD178 pKa = 4.21KK179 pKa = 11.91NQVDD183 pKa = 3.65ANFGVRR189 pKa = 11.84FAGIDD194 pKa = 3.26AAVYY198 pKa = 10.68YY199 pKa = 9.79STGTNNLKK207 pKa = 10.05QDD209 pKa = 3.33NNAYY213 pKa = 9.63ILQAMYY219 pKa = 10.56NADD222 pKa = 3.69AFKK225 pKa = 11.14VGAFYY230 pKa = 11.22SNEE233 pKa = 4.05TIEE236 pKa = 4.34NSNGSDD242 pKa = 3.69LSDD245 pKa = 3.73LNHH248 pKa = 6.41YY249 pKa = 8.88GVSGNYY255 pKa = 10.35YY256 pKa = 8.33MDD258 pKa = 3.05AWNFGLGWGAEE269 pKa = 3.97KK270 pKa = 11.12DD271 pKa = 3.64NLTSNNDD278 pKa = 3.39RR279 pKa = 11.84NDD281 pKa = 3.07YY282 pKa = 8.53YY283 pKa = 11.88ANVAYY288 pKa = 10.27AFTSNIQSYY297 pKa = 9.65VEE299 pKa = 3.9VGYY302 pKa = 10.91SDD304 pKa = 4.83EE305 pKa = 5.21DD306 pKa = 4.04DD307 pKa = 4.68SEE309 pKa = 4.34VGYY312 pKa = 10.56VVGMEE317 pKa = 4.24VVFF320 pKa = 4.62
Molecular weight: 34.65 kDa
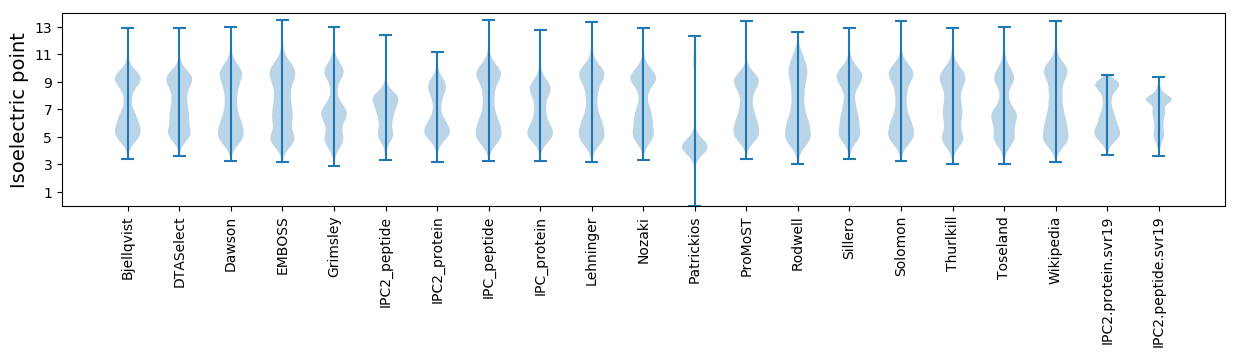
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4UI09|M4UI09_9GAMM Met regulon regulatory protein MetJ OS=Psychromonas sp. CNPT3 OX=314282 GN=PCNPT3_00390 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84SHH16 pKa = 6.27GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84SHH16 pKa = 6.27GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.2NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
852767 |
28 |
3350 |
330.8 |
36.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.087 ± 0.052 | 1.215 ± 0.02 |
5.357 ± 0.051 | 5.601 ± 0.062 |
4.398 ± 0.044 | 6.179 ± 0.047 |
2.243 ± 0.025 | 7.587 ± 0.046 |
6.503 ± 0.047 | 11.035 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.567 ± 0.027 | 4.682 ± 0.041 |
3.427 ± 0.025 | 4.562 ± 0.04 |
4.007 ± 0.037 | 6.909 ± 0.042 |
5.182 ± 0.047 | 6.259 ± 0.04 |
1.047 ± 0.019 | 3.154 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
