
Vibrio phage K05K4_VK05K4_1
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
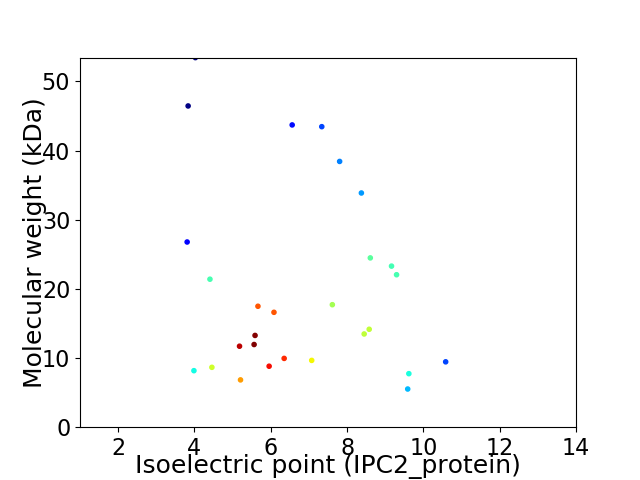
Virtual 2D-PAGE plot for 28 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6UGC1|A0A1W6UGC1_9VIRU RstB protein OS=Vibrio phage K05K4_VK05K4_1 OX=1912326 GN=rstB _3 PE=4 SV=1
MM1 pKa = 6.8NTCFLDD7 pKa = 3.66TGISTHH13 pKa = 5.61AHH15 pKa = 5.7FEE17 pKa = 3.92KK18 pKa = 10.3TPYY21 pKa = 10.84SNARR25 pKa = 11.84YY26 pKa = 8.73PYY28 pKa = 9.25QTVCDD33 pKa = 3.76NGLGLGYY40 pKa = 10.63YY41 pKa = 7.75EE42 pKa = 4.43VRR44 pKa = 11.84CPEE47 pKa = 3.81NSEE50 pKa = 4.27FDD52 pKa = 3.99PSTLRR57 pKa = 11.84CKK59 pKa = 10.33SVCEE63 pKa = 4.04YY64 pKa = 11.13GKK66 pKa = 10.26NPDD69 pKa = 3.94GTCMDD74 pKa = 3.88ACQFKK79 pKa = 10.92KK80 pKa = 10.76SIDD83 pKa = 4.02EE84 pKa = 4.15IKK86 pKa = 10.48SLQWLAYY93 pKa = 9.88VYY95 pKa = 10.64GEE97 pKa = 4.11QVTGSCYY104 pKa = 10.68GDD106 pKa = 3.56YY107 pKa = 11.09GATRR111 pKa = 11.84CEE113 pKa = 4.25LEE115 pKa = 4.18RR116 pKa = 11.84TPSDD120 pKa = 3.21STLCTGVDD128 pKa = 3.27SGQWTQTPSVMVTFNSQATQCEE150 pKa = 4.55GGTLFWGKK158 pKa = 10.11DD159 pKa = 3.62GPDD162 pKa = 3.31TPIIPDD168 pKa = 4.37DD169 pKa = 5.14PIHH172 pKa = 7.61DD173 pKa = 4.88PDD175 pKa = 5.88DD176 pKa = 3.89PTGDD180 pKa = 3.64IEE182 pKa = 5.54DD183 pKa = 4.5PSILPDD189 pKa = 3.61GSTNTVNPPDD199 pKa = 4.02TDD201 pKa = 3.65SEE203 pKa = 4.63PDD205 pKa = 3.48VEE207 pKa = 5.55EE208 pKa = 5.22PDD210 pKa = 3.63TDD212 pKa = 3.49EE213 pKa = 4.45STDD216 pKa = 3.64TAVLKK221 pKa = 10.91AITGMNKK228 pKa = 10.0DD229 pKa = 3.38VNKK232 pKa = 10.54ALNDD236 pKa = 3.47MNIDD240 pKa = 3.4INQANADD247 pKa = 3.75VQNQIIALNASMVTNTQAIQKK268 pKa = 7.84QQINDD273 pKa = 3.14NKK275 pKa = 10.37IYY277 pKa = 10.49EE278 pKa = 4.16NTKK281 pKa = 10.95ALIQQANADD290 pKa = 3.49ITTAMNKK297 pKa = 7.52NTNAVNGVGDD307 pKa = 4.45DD308 pKa = 3.72VEE310 pKa = 5.55KK311 pKa = 10.74IAGAMDD317 pKa = 5.45GIAEE321 pKa = 4.26DD322 pKa = 3.65VSGISDD328 pKa = 3.9TLDD331 pKa = 4.1GIANTDD337 pKa = 3.08TSGAGTGGTCIEE349 pKa = 4.43SQSCTGFYY357 pKa = 10.69EE358 pKa = 4.04SGYY361 pKa = 9.68PDD363 pKa = 3.62GLGGLVSGQLDD374 pKa = 3.73DD375 pKa = 5.58LKK377 pKa = 11.35HH378 pKa = 4.84NTIDD382 pKa = 3.42NFVNSFGDD390 pKa = 3.73LDD392 pKa = 4.02LSSAKK397 pKa = 10.1RR398 pKa = 11.84PSFVLPVPFFGDD410 pKa = 3.76FSFEE414 pKa = 4.0EE415 pKa = 4.36QISFDD420 pKa = 3.97LLFGLFVRR428 pKa = 11.84CSSS431 pKa = 3.36
MM1 pKa = 6.8NTCFLDD7 pKa = 3.66TGISTHH13 pKa = 5.61AHH15 pKa = 5.7FEE17 pKa = 3.92KK18 pKa = 10.3TPYY21 pKa = 10.84SNARR25 pKa = 11.84YY26 pKa = 8.73PYY28 pKa = 9.25QTVCDD33 pKa = 3.76NGLGLGYY40 pKa = 10.63YY41 pKa = 7.75EE42 pKa = 4.43VRR44 pKa = 11.84CPEE47 pKa = 3.81NSEE50 pKa = 4.27FDD52 pKa = 3.99PSTLRR57 pKa = 11.84CKK59 pKa = 10.33SVCEE63 pKa = 4.04YY64 pKa = 11.13GKK66 pKa = 10.26NPDD69 pKa = 3.94GTCMDD74 pKa = 3.88ACQFKK79 pKa = 10.92KK80 pKa = 10.76SIDD83 pKa = 4.02EE84 pKa = 4.15IKK86 pKa = 10.48SLQWLAYY93 pKa = 9.88VYY95 pKa = 10.64GEE97 pKa = 4.11QVTGSCYY104 pKa = 10.68GDD106 pKa = 3.56YY107 pKa = 11.09GATRR111 pKa = 11.84CEE113 pKa = 4.25LEE115 pKa = 4.18RR116 pKa = 11.84TPSDD120 pKa = 3.21STLCTGVDD128 pKa = 3.27SGQWTQTPSVMVTFNSQATQCEE150 pKa = 4.55GGTLFWGKK158 pKa = 10.11DD159 pKa = 3.62GPDD162 pKa = 3.31TPIIPDD168 pKa = 4.37DD169 pKa = 5.14PIHH172 pKa = 7.61DD173 pKa = 4.88PDD175 pKa = 5.88DD176 pKa = 3.89PTGDD180 pKa = 3.64IEE182 pKa = 5.54DD183 pKa = 4.5PSILPDD189 pKa = 3.61GSTNTVNPPDD199 pKa = 4.02TDD201 pKa = 3.65SEE203 pKa = 4.63PDD205 pKa = 3.48VEE207 pKa = 5.55EE208 pKa = 5.22PDD210 pKa = 3.63TDD212 pKa = 3.49EE213 pKa = 4.45STDD216 pKa = 3.64TAVLKK221 pKa = 10.91AITGMNKK228 pKa = 10.0DD229 pKa = 3.38VNKK232 pKa = 10.54ALNDD236 pKa = 3.47MNIDD240 pKa = 3.4INQANADD247 pKa = 3.75VQNQIIALNASMVTNTQAIQKK268 pKa = 7.84QQINDD273 pKa = 3.14NKK275 pKa = 10.37IYY277 pKa = 10.49EE278 pKa = 4.16NTKK281 pKa = 10.95ALIQQANADD290 pKa = 3.49ITTAMNKK297 pKa = 7.52NTNAVNGVGDD307 pKa = 4.45DD308 pKa = 3.72VEE310 pKa = 5.55KK311 pKa = 10.74IAGAMDD317 pKa = 5.45GIAEE321 pKa = 4.26DD322 pKa = 3.65VSGISDD328 pKa = 3.9TLDD331 pKa = 4.1GIANTDD337 pKa = 3.08TSGAGTGGTCIEE349 pKa = 4.43SQSCTGFYY357 pKa = 10.69EE358 pKa = 4.04SGYY361 pKa = 9.68PDD363 pKa = 3.62GLGGLVSGQLDD374 pKa = 3.73DD375 pKa = 5.58LKK377 pKa = 11.35HH378 pKa = 4.84NTIDD382 pKa = 3.42NFVNSFGDD390 pKa = 3.73LDD392 pKa = 4.02LSSAKK397 pKa = 10.1RR398 pKa = 11.84PSFVLPVPFFGDD410 pKa = 3.76FSFEE414 pKa = 4.0EE415 pKa = 4.36QISFDD420 pKa = 3.97LLFGLFVRR428 pKa = 11.84CSSS431 pKa = 3.36
Molecular weight: 46.45 kDa
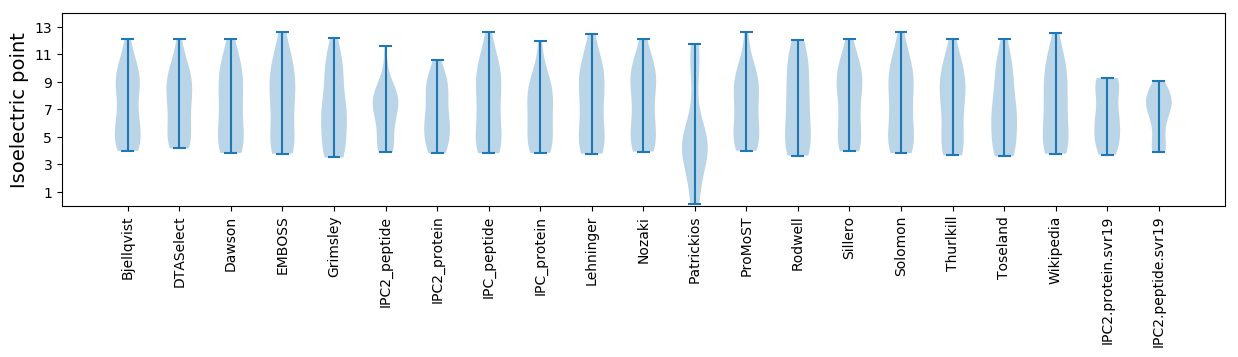
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6UFW1|A0A1W6UFW1_9VIRU Uncharacterized protein OS=Vibrio phage K05K4_VK05K4_1 OX=1912326 GN=K05K4_52540 PE=4 SV=1
MM1 pKa = 7.41AKK3 pKa = 9.95SVFVLGMDD11 pKa = 3.58ITWNSARR18 pKa = 11.84GDD20 pKa = 3.84SAQLNVSRR28 pKa = 11.84PLRR31 pKa = 11.84EE32 pKa = 3.83INSEE36 pKa = 3.91KK37 pKa = 10.33FKK39 pKa = 11.11RR40 pKa = 11.84RR41 pKa = 11.84ALSVNPVMM49 pKa = 5.81
MM1 pKa = 7.41AKK3 pKa = 9.95SVFVLGMDD11 pKa = 3.58ITWNSARR18 pKa = 11.84GDD20 pKa = 3.84SAQLNVSRR28 pKa = 11.84PLRR31 pKa = 11.84EE32 pKa = 3.83INSEE36 pKa = 3.91KK37 pKa = 10.33FKK39 pKa = 11.11RR40 pKa = 11.84RR41 pKa = 11.84ALSVNPVMM49 pKa = 5.81
Molecular weight: 5.52 kDa
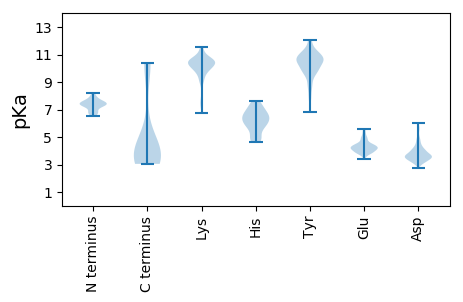
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5065 |
49 |
491 |
180.9 |
20.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.029 ± 0.405 | 2.113 ± 0.252 |
6.693 ± 0.55 | 5.429 ± 0.228 |
4.423 ± 0.187 | 6.752 ± 0.392 |
1.856 ± 0.203 | 5.884 ± 0.305 |
6.199 ± 0.41 | 8.687 ± 0.549 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.567 ± 0.276 | 4.877 ± 0.404 |
4.284 ± 0.238 | 4.166 ± 0.299 |
4.837 ± 0.477 | 6.614 ± 0.305 |
6.338 ± 0.47 | 6.12 ± 0.357 |
1.461 ± 0.159 | 3.672 ± 0.307 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
